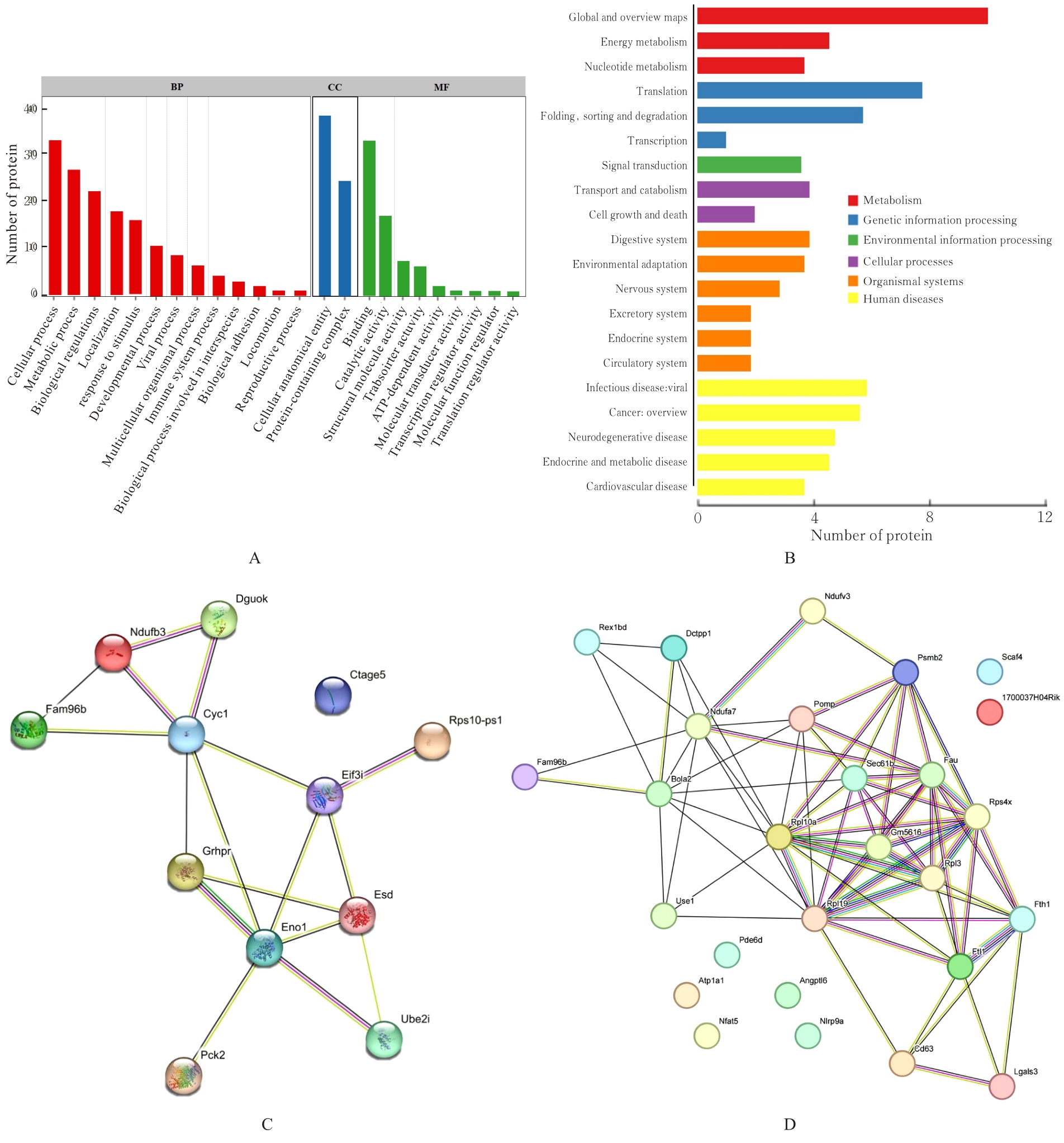
| 1 |
DAY J M. The diversity of the orthoreoviruses: molecular taxonomy and phylogentic divides[J]. Infect Genet Evol, 2009, 9(4): 390-400.
|
| 2 |
GARD G, COMPANS R W. Structure and cytopathic effects of Nelson Bay virus[J]. J Virol, 1970, 6(1): 100-106.
|
| 3 |
KUANG G P, XU Z Q, WANG J, et al. Nelson bay reovirus isolated from bats and blood-sucking arthropods collected in Yunnan Province, China[J]. Microbiol Spectr, 2023, 11(4): e0512222.
|
| 4 |
MERTENS P. The dsRNA viruses[J]. Virus Res, 2004, 101(1): 3-13.
|
| 5 |
BENAVENTE J, MARTÍNEZ-COSTAS J. Avian reovirus: structure and biology[J]. Virus Res, 2007, 123(2): 105-119.
|
| 6 |
HOTANI T, TACHIBANA M, MIZUGUCHI H, et al. Reovirus double-stranded RNA genomes and polyⅠ: C induce down-regulation of hypoxia-inducible factor 1α[J]. Biochem Biophys Res Commun, 2015, 460(4): 1041-1046.
|
| 7 |
DUNCAN R. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal[J]. Virology, 1999, 260(2): 316-328.
|
| 8 |
ZAMORA P F, HU L Y, KNOWLTON J J, et al. Reovirus nonstructural protein σNS acts as an RNA stability factor promoting viral genome replication[J]. J Virol, 2018, 92(15): e00563-18.
|
| 9 |
TENORIO R, FERNÁNDEZ DE CASTRO I, KNOWLTON J J, et al. Function, architecture, and biogenesis of reovirus replication neoorganelles[J]. Viruses, 2019, 11(3): 288.
|
| 10 |
TENORIO R, FERNÁNDEZ DE CASTRO I, KNOWLTON J J, et al. Reovirus σNS and μNS proteins remodel the endoplasmic reticulum to build replication neo-organelles[J]. mBio, 2018, 9(4): e01253-18.
|
| 11 |
LEE C H, RAGHUNATHAN K, TAYLOR G M, et al. Reovirus nonstructural protein σNS recruits viral RNA to replication organelles[J]. mBio, 2021, 12(4): e0140821.
|
| 12 |
GORAL M I, MOCHOW-GRUNDY M, DERMODY T S. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation[J]. Virology, 1996, 216(1): 265-271.
|
| 13 |
KUMAR R, MEHTA D, MISHRA N, et al. Role of host-mediated post-translational modifications (PTMs) in RNA virus pathogenesis[J]. Int J Mol Sci, 2020, 22(1): 323.
|
| 14 |
MOK L, WYNNE J W, GRIMLEY S, et al. Mouse fibroblast L929 cells are less permissive to infection by Nelson Bay orthoreovirus compared to other mammalian cell lines[J]. J Gen Virol, 2015, 96(Pt 7): 1787-1794.
|
| 15 |
MOK L, WYNNE J W, TACHEDJIAN M, et al. Proteomics informed by transcriptomics for characterising differential cellular susceptibility to Nelson Bay orthoreovirus infection[J]. BMC Genomics, 2017, 18(1): 615.
|
| 16 |
CHIEN C T, BARTEL P L, STERNGLANZ R, et al. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest[J]. Proc Natl Acad Sci U S A, 1991, 88(21): 9578-9582.
|
| 17 |
RAO B C, LI J H, REN T, et al. RPL19 is a prognostic biomarker and promotes tumor progression in hepatocellular carcinoma[J]. Front Cell Dev Biol, 2021, 9: 686547.
|
| 18 |
PETROVA E, GRACIAS S, BEAUCLAIR G, et al. Uncovering flavivirus host dependency factors through a genome-wide gain-of-function screen[J]. Viruses, 2019, 11(1): 68.
|
| 19 |
PAN W, SONG D G, HE W Q, et al. EIF3i affects vesicular stomatitis virus growth by interacting with matrix protein[J]. Vet Microbiol, 2017, 212: 59-66.
|
| 20 |
KOMAROVA A V, REAL E, BORMAN A M, et al. Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis[J]. Nucleic Acids Res, 2007, 35(5): 1522-1532.
|
| 21 |
YIN X J, HONG W, TIAN F J, et al. Proteomic analysis of decidua in patients with recurrent pregnancy loss (RPL) reveals mitochondrial oxidative stress dysfunction[J]. Clin Proteomics, 2021, 18(1): 9.
|
| 22 |
GUARNIERI J W, DYBAS J M, FAZELINIA H, et al. Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts[J]. Sci Transl Med, 2023, 15(708): eabq1533.
|
| 23 |
MUHAMMAD J S, ELGHAZALI G, SHAFARIN J, et al. SARS-CoV-2-induced hypomethylation of the ferritin heavy chain (FTH1) gene underlies serum hyperferritinemia in severe COVID-19 patients[J]. Biochem Biophys Res Commun, 2022, 631: 138-145.
|
| 24 |
OHTA K, SAKA N, NISHIO M. Human parainfluenza virus type 2 V protein modulates iron homeostasis[J]. J Virol, 2021, 95(6): e01861.
|
 )
)