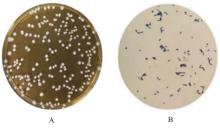
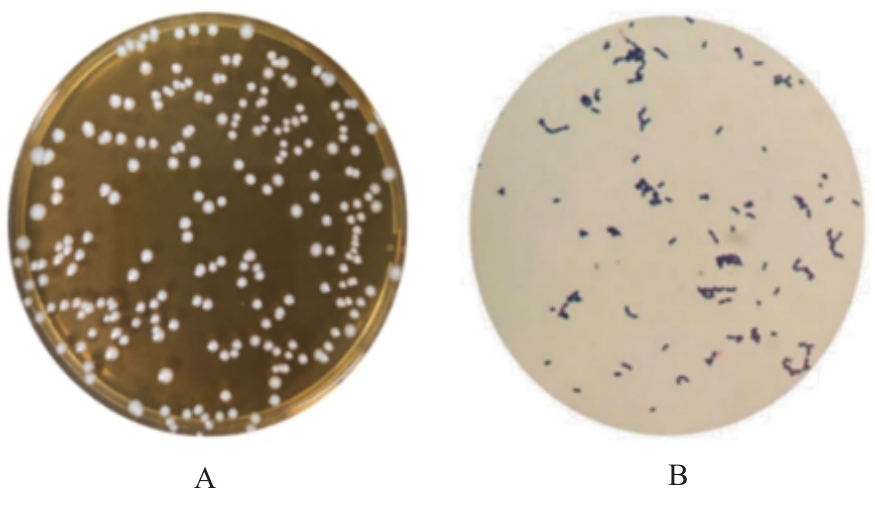
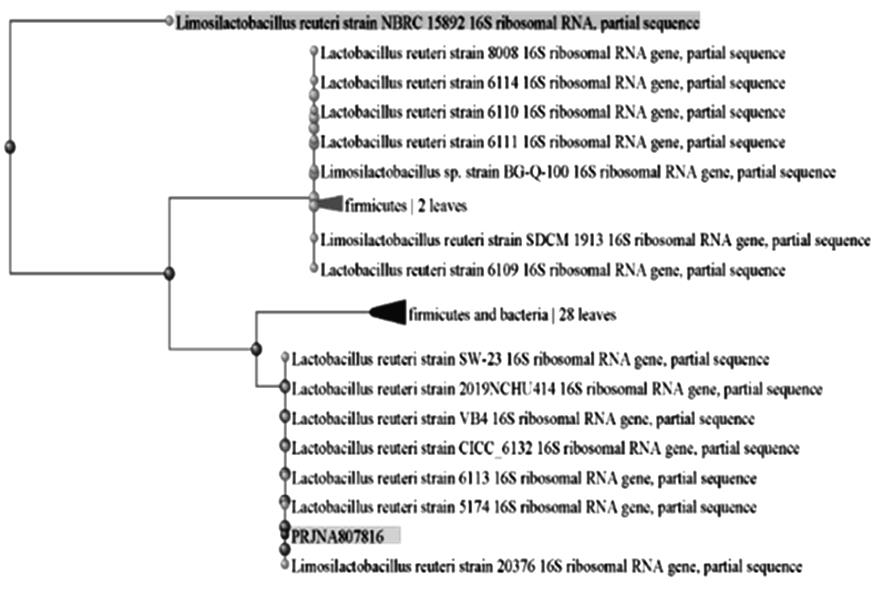
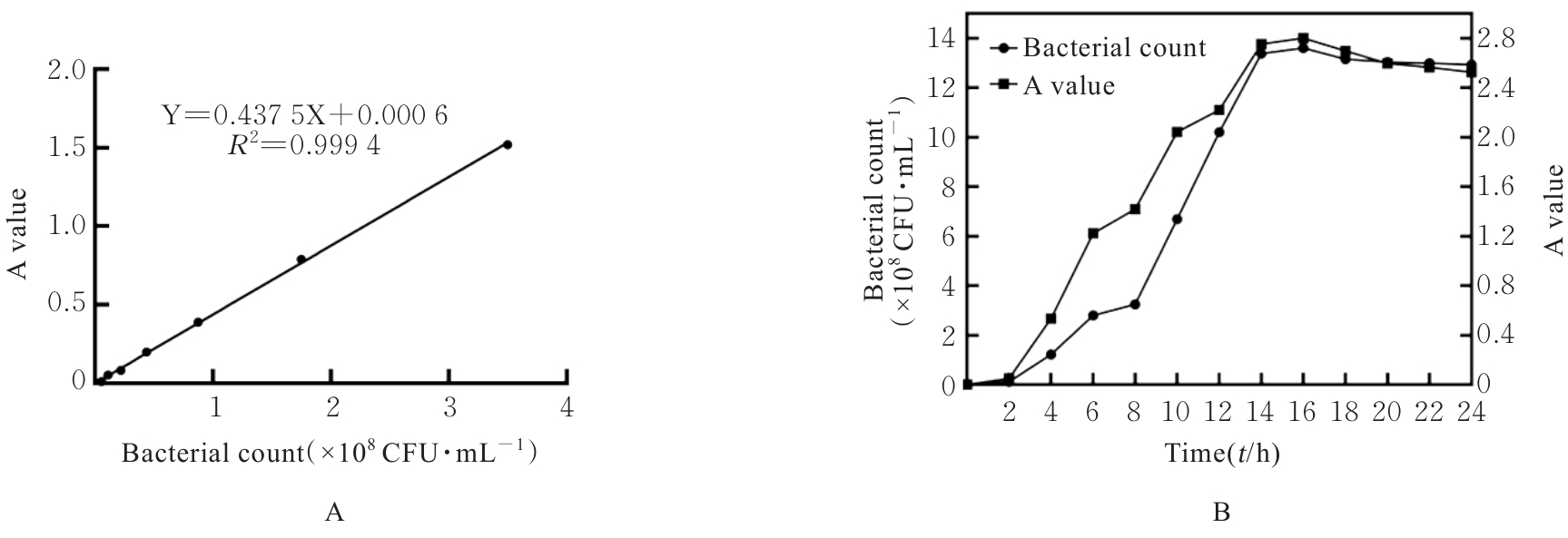
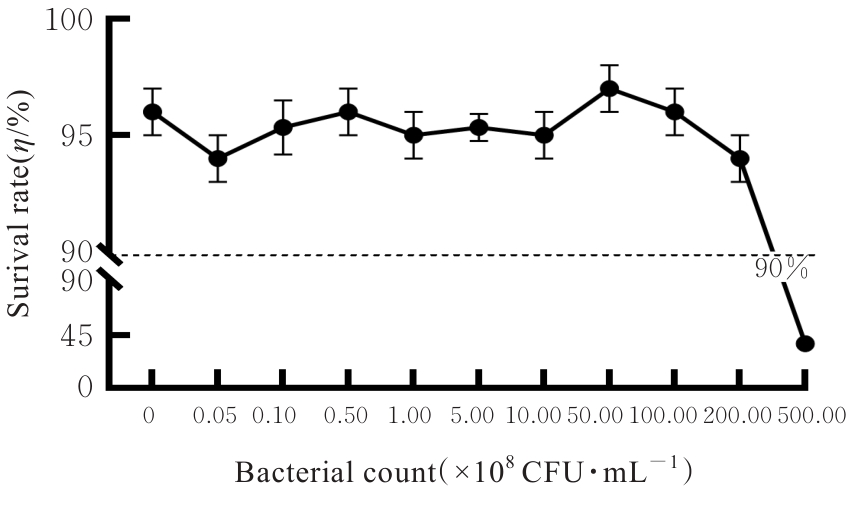
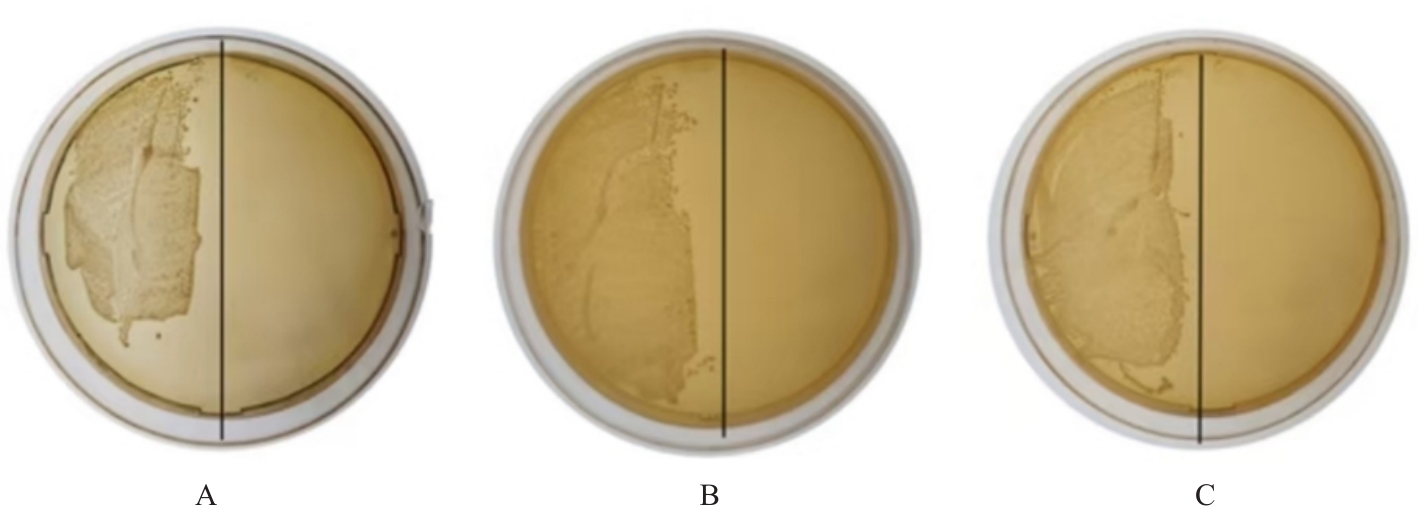
| 1 |
MU Q H, TAVELLA V J, LUO X M. Role of Lactobacillus reuteri in human health and diseases[J]. Front Microbiol, 2018, 9: 757.
|
| 2 |
GUARINO A, GUANDALINI S, VECCHIO A L. Probiotics for prevention and treatment of diarrhea[J]. J Clin Gastroenterol, 2015, 49(): S37-S45.
|
| 3 |
WU H Q, XIE S, MIAO J F, et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa[J]. Gut Microbes, 2020, 11(4): 997-1014.
|
| 4 |
AL-HADIDI A, NAVARRO J, GOODMAN S D, et al. Lactobacillus reuteri in its biofilm state improves protection from experimental necrotizing enterocolitis[J]. Nutrients, 2021, 13(3): 918.
|
| 5 |
ZHOU Q H, WU F T, CHEN S W, et al. Lactobacillus reuteri improves function of the intestinal barrier in rats with acute liver failure through Nrf-2/HO-1 pathway[J]. Nutrition, 2022, 99/100: 111673.
|
| 6 |
PENG Y J, MA Y Z, LUO Z C, et al. Lactobacillus reuteri in digestive system diseases: focus on clinical trials and mechanisms[J]. Front Cell Infect Microbiol, 2023, 13: 1254198.
|
| 7 |
AMOROSO C, PERILLO F, STRATI F, et al. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation[J]. Cells, 2020, 9(5): 1234.
|
| 8 |
WILKINS T, SEQUOIA J. Probiotics for gastrointestinal conditions: a summary of the evidence[J]. Am Fam Physician, 2017, 96(3): 170-178.
|
| 9 |
SUN X Q, KONG J, ZHU S T, et al. A systematic review and meta-analysis: the therapeutic and preventive effect of Lactobacillus reuteri DSM 17,938 addition in children with diarrhea[J]. BMC Gastroenterol, 2023, 23(1): 141.
|
| 10 |
AZIZ A B, ALI M, BASUNIA A H, et al. Impact of vaccination on the risk factors for acute rotavirus diarrhea: an analysis of the data of a cluster randomized trial conducted in a rural area of Bangladesh[J]. Vaccine, 2020, 38(9): 2190-2197.
|
| 11 |
ZHAO W, YU M L, TAO X L, et al. Analysis of the intestinal microbial community altered during rotavirus infection in suckling mice[J]. Virol J, 2021, 18(1): 254.
|
| 12 |
VALENCIA-RODRÍGUEZ A, AQUINO-MATUS J, VERA-BARAJAS A, et al. New therapeutic options for bile acid malabsorption diarrhea[J]. Ann Transl Med, 2019, 7(22): 695.
|
| 13 |
WIRUSANTI N I, BALDRIDGE M T, HARRIS V C. Microbiota regulation of viral infections through interferon signaling[J]. Trends Microbiol, 2022, 30(8): 778-792.
|
| 14 |
KUSS S K, BEST G T, ETHEREDGE C A, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis[J]. Science, 2011, 334(6053): 249-252.
|
| 15 |
GRAU K R, ZHU S, PETERSON S T, et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type Ⅲ interferon[J]. Nat Microbiol, 2020, 5(1): 84-92.
|
| 16 |
KNIPPING K, MCNEAL M M, CRIENEN A, et al. A gastrointestinal rotavirus infection mouse model for immune modulation studies[J]. Virol J, 2011, 8: 109.
|
| 17 |
RIGO-ADROVER M D M, KNIPPING K, GARSSEN J, et al. Prevention of rotavirus diarrhea in suckling rats by a specific fermented milk concentrate with prebiotic mixture[J]. Nutrients, 2019, 11(1): 189.
|
| 18 |
KARST S M. The influence of commensal bacteria on infection with enteric viruses[J]. Nat Rev Microbiol, 2016, 14(4): 197-204.
|
| 19 |
CHATTHA K S, VLASOVA A N, KANDASAMY S, et al. Divergent immunomodulating effects of probiotics on T cell responses to oral attenuated human rotavirus vaccine and virulent human rotavirus infection in a neonatal gnotobiotic piglet disease model[J]. J Immunol, 2013, 191(5): 2446-2456.
|
| 20 |
VLASOVA A N, CHATTHA K S, KANDASAMY S, et al. Lactobacilli and bifidobacteria promote immune homeostasis by modulating innate immune responses to human rotavirus in neonatal gnotobiotic pigs[J]. PLoS One, 2013, 8(10): e76962.
|
| 21 |
SHI Z D, ZOU J, ZHANG Z, et al. Segmented filamentous bacteria prevent and cure rotavirus infection[J]. Cell, 2019, 179(3): 644-658.e13.
|
| 22 |
PAPARO L, TRIPODI L, BRUNO C, et al. Protective action of Bacillus clausii probiotic strains in an in vitro model of Rotavirus infection[J]. Sci Rep, 2020, 10(1): 12636.
|
| 23 |
FERNANDEZ-DUARTE K P, OLAYA-GALÁN N N, SALAS-CÁRDENAS S P, et al. Bifidobacterium adolescentis (DSM 20083) and Lactobacillus casei (lafti L26-DSL): probiotics able to block the in vitro adherence of rotavirus in MA104 cells[J]. Probiotics Antimicrob Proteins, 2018, 10(1): 56-63.
|
| 24 |
DOMÍNGUEZ-DÍAZ C, GARCÍA-OROZCO A, RIERA-LEAL A, et al. Microbiota and its role on viral evasion: is it with us or against us?[J]. Front Cell Infect Microbiol, 2019, 9: 256.
|
 )
)