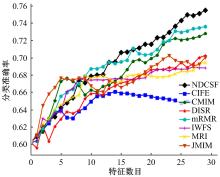
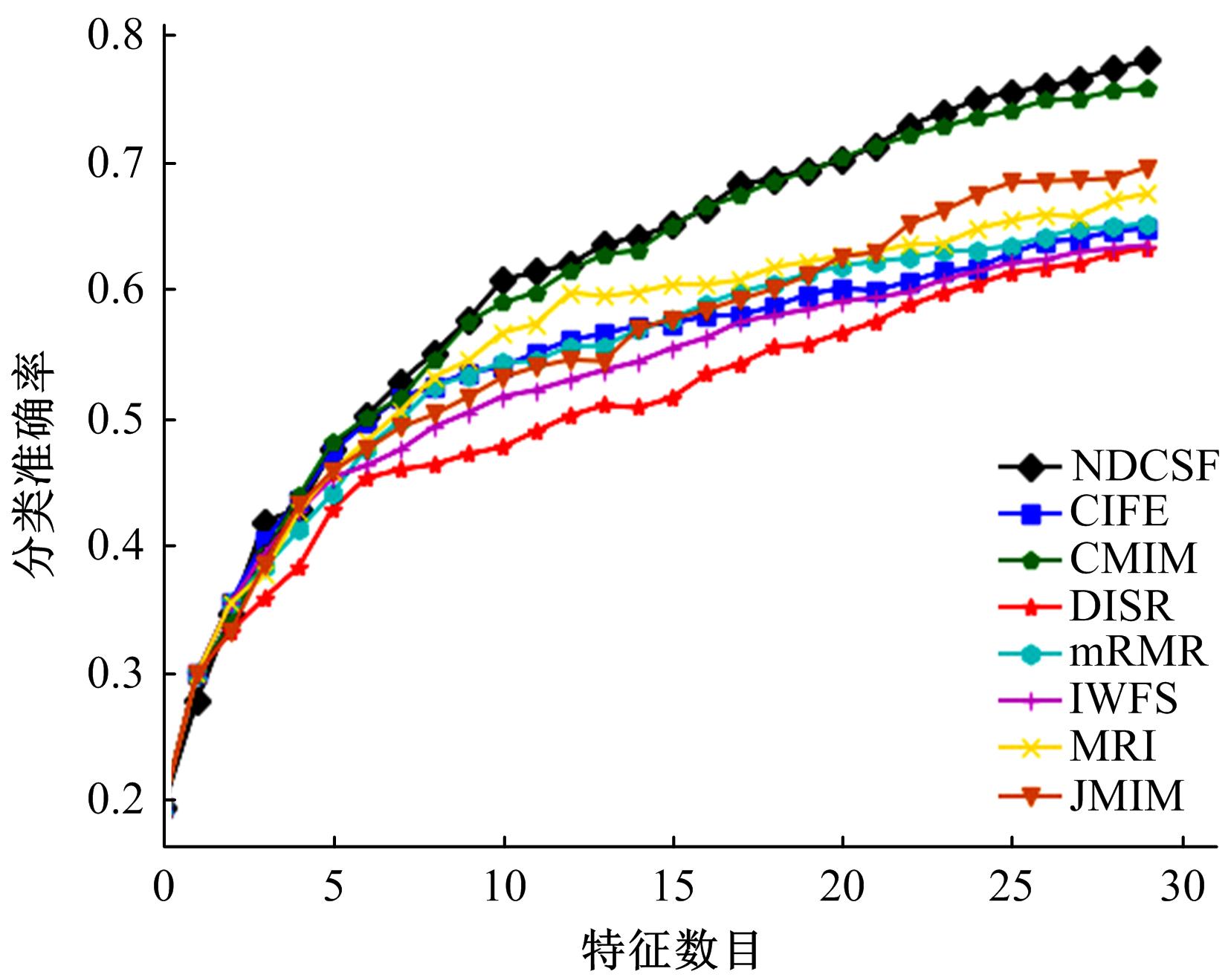
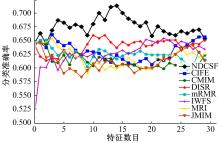
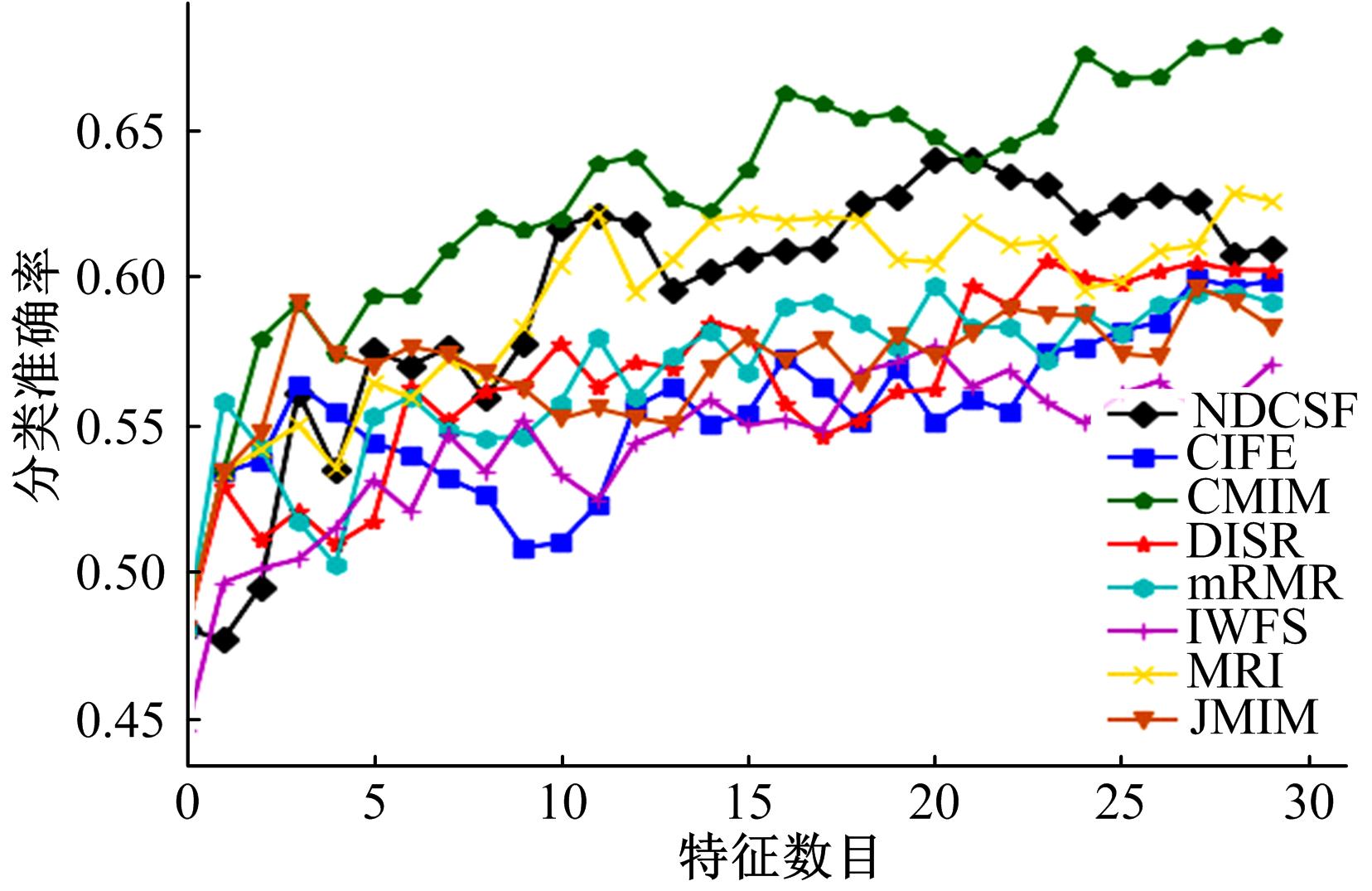
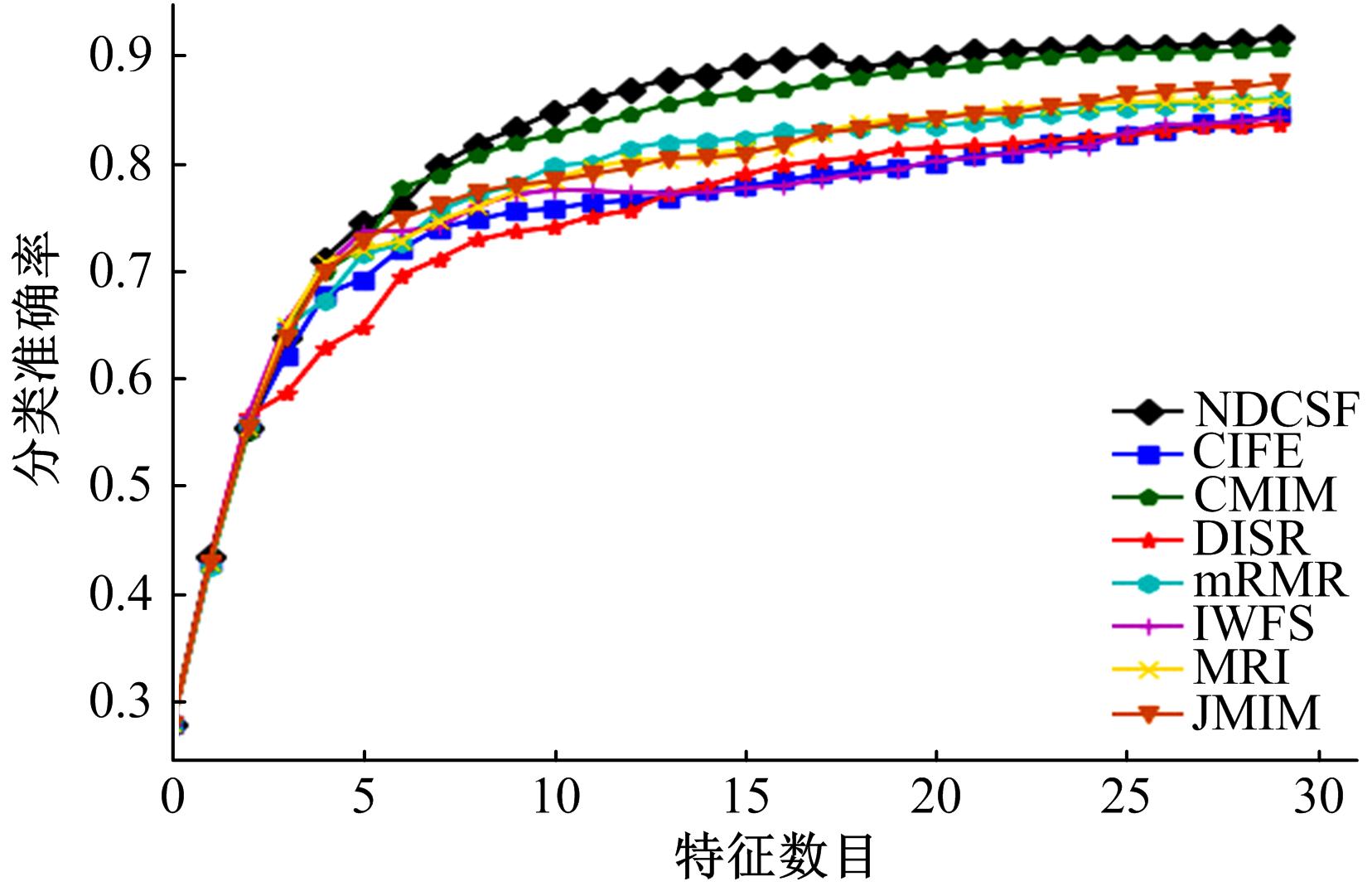
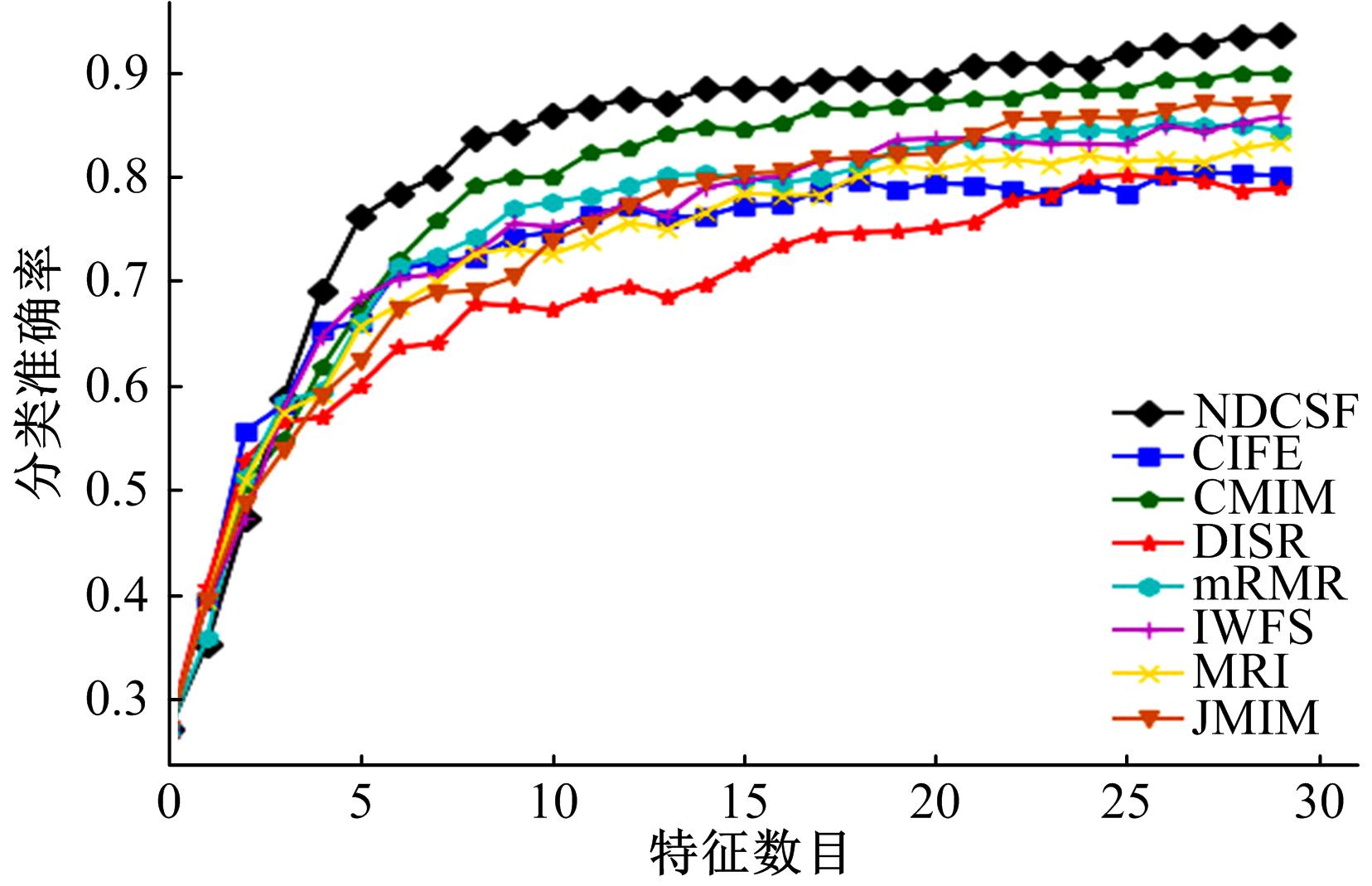
Journal of Jilin University(Engineering and Technology Edition) ›› 2019, Vol. 49 ›› Issue (4): 1293-1300.doi: 10.13229/j.cnki.jdxbgxb20180478
Nonlinear feature selection method based on dynamic change of selected features
Wan-fu GAO1( ),Ping ZHANG2,Liang HU1(
),Ping ZHANG2,Liang HU1( )
)
- 1. College of Computer Science and Technology,Jilin University,Changchun 130012, China
2. College of Software,Jilin University,Changchun 130012,China
CLC Number:
- TP301
| 1 | Gao W , Hu L , Zhang P . Class-specific mutual information variation for feature selection[J]. Pattern Recognition, 2018, 79:328-339. |
| 2 | Hu L , Gao W , Zhao K , et al . Feature selection considering two types of feature relevancy and feature interdependency[J]. Expert Systems with Applications, 2018, 93:423-434. |
| 3 | 石峰,莫忠息 .信息论基础[M].3版.武汉:武汉大学出版社,2014:14-52. |
| 4 | 赵晓群 .信息论基础及应用[M].北京:机械工业出版社,2015:27-53. |
| 5 | Lewis D D . Feature selection and feature extraction for text categorization[C]⫽The Workshop on Speech & Natural Language, Association for Computational Linguistics,Harriman, New York, 1992:212-217. |
| 6 | Battiti R . Using mutual information for selecting features in supervised neural net learning[J]. IEEE Transactions on Neural Networks, 1994, 5(4):537-550. |
| 7 | Peng H , Long F , Ding C . Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy[J]. IEEE Transactions on Pattern Analysis & Machine Intelligence, 2005, 27(8):1226-1238. |
| 8 | Lin D , Tang X . Conditional infomax learning: an integrated framework for feature extraction and fusion[C]⫽European Conference on Computer Vision, Graz, 2006: 68-82. |
| 9 | Salueña C , Avalos J B . Information-theoretic feature selection in microarray data using variable complementarity[J]. IEEE Journal of Selected Topics in Signal Processing, 2008, 2(3):261-274. |
| 10 | Zeng Z , Zhang H , Zhang R , et al . A novel feature selection method considering feature interaction[J]. Pattern Recognition, 2015, 48(8):2656-2666. |
| 11 | Fleuret F . Fast Binary Feature selection with conditional mutual information[J]. Journal of Machine Learning Research, 2004, 5(3):1531-1555. |
| 12 | Bennasar M , Hicks Y , Setchi R . Feature selection using joint mutual information maximisation[J]. Expert Systems with Applications, 2015, 42(22):8520-8532. |
| 13 | Wang J , Wei J M , Yang Z , et al . Feature selection by maximizing independent classification information[J]. IEEE Transactions on Knowledge & Data Engineering, 2017, 29(4):828-841. |
| 14 | Che J , Yang Y , Li L , et al . Maximum relevance minimum common redundancy feature selection for nonlinear data[J]. Information Sciences, 2017, 409:68-86. |
| 15 | Li J , Cheng K , Wang S , et al . Feature selection: a data perspective[J]. Acm Computing Surveys, 2016, 50(6). |
| 16 | Lichman M . UCI machine learning repository[DB/OL].[2018-05-15].. |
| [1] | Dan⁃tong OUYANG,Jun XIAO,Yu⁃xin YE. Distant supervision for relation extraction with weakconstraints of entity pairs [J]. Journal of Jilin University(Engineering and Technology Edition), 2019, 49(3): 912-919. |
| [2] | DONG Sa, LIU Da-you, OUYANG Ruo-chuan, ZHU Yun-gang, LI Li-na. Logistic regression classification in networked data with heterophily based on second-order Markov assumption [J]. Journal of Jilin University(Engineering and Technology Edition), 2018, 48(5): 1571-1577. |
| [3] | GU Hai-jun, TIAN Ya-qian, CUI Ying. Intelligent interactive agent for home service [J]. Journal of Jilin University(Engineering and Technology Edition), 2018, 48(5): 1578-1585. |
| [4] | WANG Xu, OUYANG Ji-hong, CHEN Gui-fen. Measurement of graph similarity based on vertical dimension sequence dynamic time warping method [J]. 吉林大学学报(工学版), 2018, 48(4): 1199-1205. |
| [5] | ZHANG Hao, ZHAN Meng-ping, GUO Liu-xiang, LI Zhi, LIU Yuan-ning, ZHANG Chun-he, CHANG Hao-wu, WANG Zhi-qiang. Human exogenous plant miRNA cross-kingdom regulatory modeling based on high-throughout data [J]. 吉林大学学报(工学版), 2018, 48(4): 1206-1213. |
| [6] | LI Xiong-fei, FENG Ting-ting, LUO Shi, ZHANG Xiao-li. Automatic music composition algorithm based on recurrent neural network [J]. 吉林大学学报(工学版), 2018, 48(3): 866-873. |
| [7] | HUANG Lan, JI Lin-ying, YAO Gang, ZHAI Rui-feng, BAI Tian. Construction of disease-symptom semantic net for misdiagnosis prompt [J]. 吉林大学学报(工学版), 2018, 48(3): 859-865. |
| [8] | LIU Jie, ZHANG Ping, GAO Wan-fu. Feature selection method based on conditional relevance [J]. 吉林大学学报(工学版), 2018, 48(3): 874-881. |
| [9] | CHEN Tao, CUI Yue-han, GUO Li-min. Improved algorithm of multiple signal classification for single snapshot [J]. 吉林大学学报(工学版), 2018, 48(3): 952-956. |
| [10] | CHEN Mian-shu, SU Yue, SANG Ai-jun, LI Pei-peng. Image classification methods based on space vector model [J]. 吉林大学学报(工学版), 2018, 48(3): 943-951. |
| [11] | LIU Xue-juan, YUAN Jia-bin, XU Juan, DUAN Bo-jia. Quantum k-means algorithm [J]. 吉林大学学报(工学版), 2018, 48(2): 539-544. |
| [12] | WANG Xu, OUYANG Ji-hong, CHEN Gui-fen. Heuristic algorithm of all common subsequences of multiple sequences for measuring multiple graphs similarity [J]. 吉林大学学报(工学版), 2018, 48(2): 526-532. |
| [13] | YANG Xin, XIA Si-jun, LIU Dong-xue, FEI Shu-min, HU Yin-ji. Target tracking based on improved accelerated gradient under tracking-learning-detection framework [J]. 吉林大学学报(工学版), 2018, 48(2): 533-538. |
| [14] | YANG Hong-yu, XU Jin. An Android malware static detection model [J]. 吉林大学学报(工学版), 2018, 48(2): 564-570. |
| [15] | LI Jia-fei, SUN Xiao-yu. Clustering method for uncertain data based on spectral decomposition [J]. 吉林大学学报(工学版), 2017, 47(5): 1604-1611. |
|
||