吉林大学学报(医学版) ›› 2022, Vol. 48 ›› Issue (3): 801-808.doi: 10.13481/j.1671-587X.20220331
• 方法学 • 上一篇
真伪熊胆粉物种基源分子生物学鉴定方法的建立及评价
- 北华大学医学技术学院分子生物学教研室,吉林 吉林 132013
Establishment of molecular biological identification method for origin of true and false bear bile powder species and its evaluation
Yuan ZHAO,Huijian JIA,Shunjia SONG,Qi LI,Xuechao SHAO,Jinxia AI,Liyuan SUN( )
)
- Department of Molecular Biology,School of Medical Technology,Beihua University,Jilin Jilin,132013,China
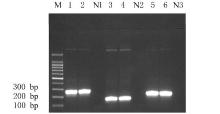
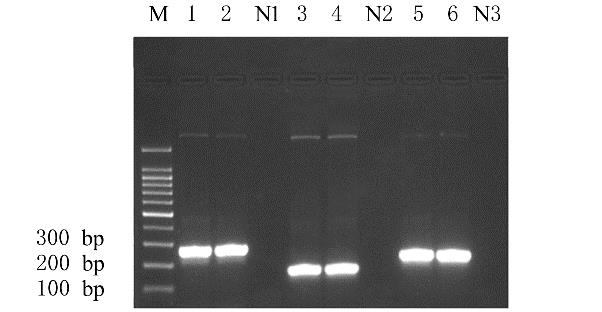
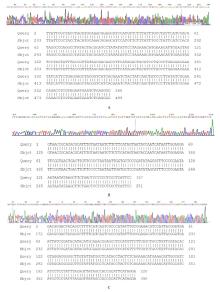
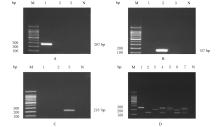
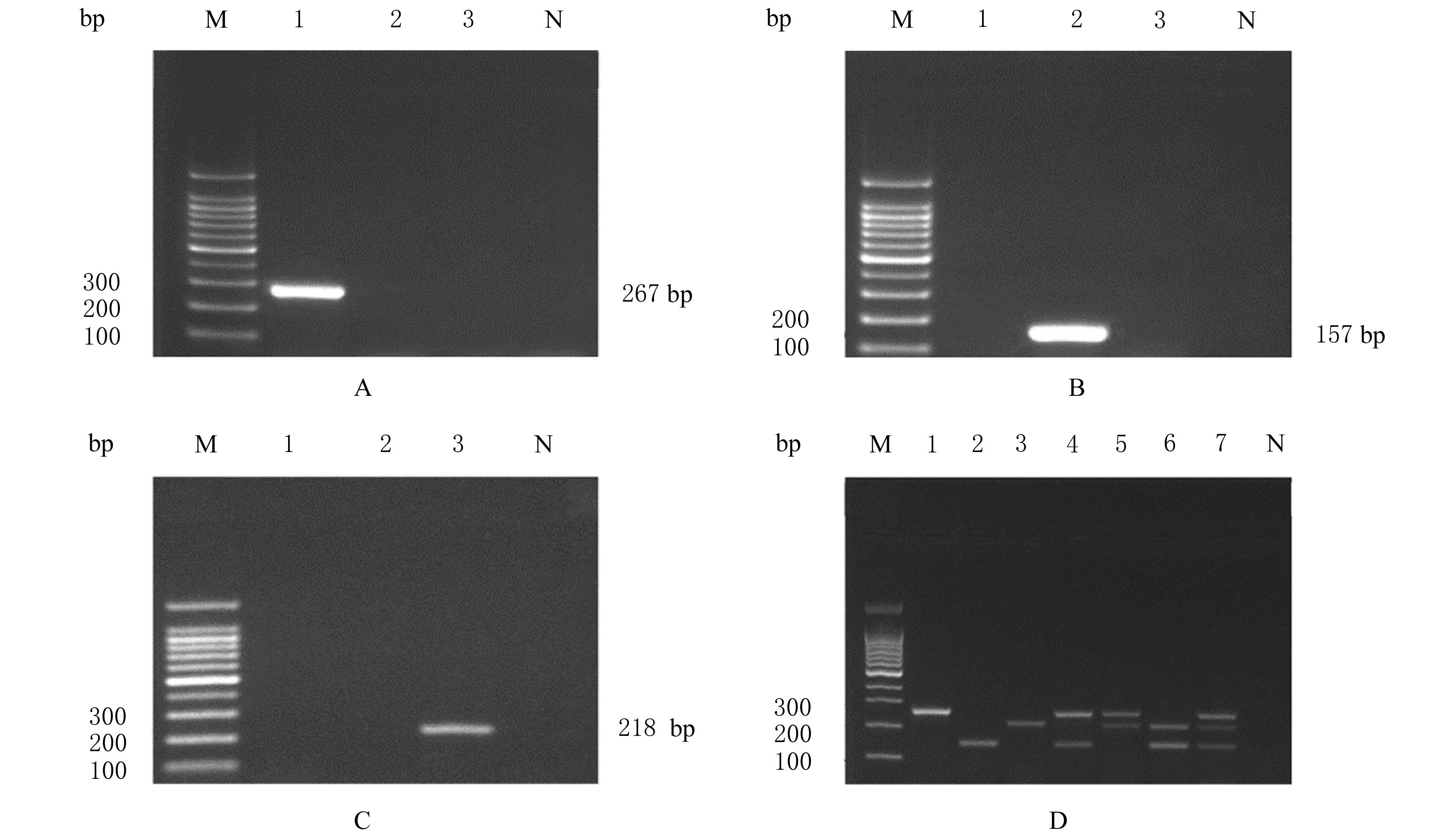
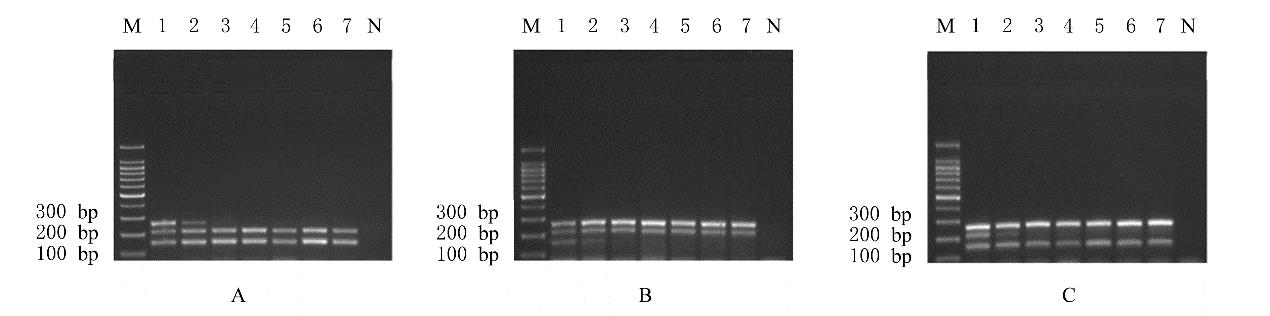
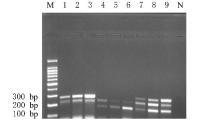
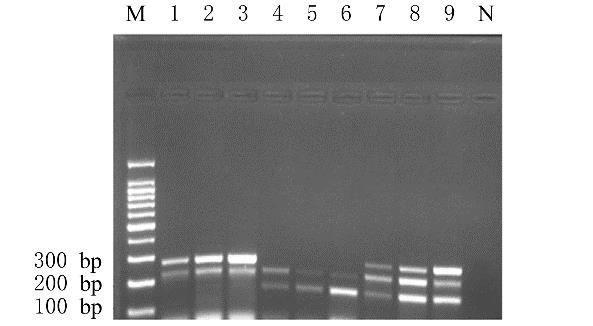
摘要: 基于多重聚合酶链式反应(PCR)技术,建立并评价一种可同时快速特异鉴别熊胆粉生物来源的方法,为鉴别人工模拟混合掺假样品提供依据。 利用猪、牛和熊线粒体细胞色素b基因,SDS-蛋白酶K裂解法(SDS-PK)提取胆类DNA,Primer Premier 5.0软件设计可扩增不同大小DNA片段且无交叉反应的物种特异性引物,特异性扩增后,将扩增条带克隆后测序,与GenBank数据库已登记序列比对。建立并优化多重PCR,评价其特异性和灵敏度,并采用该方法鉴别27份模拟混合掺假样品。 建立的猪、牛和熊胆粉DNA提取方法可于2 h之内完成操作,DNA纯度分别为2.04、1.69和1.70,浓度分别为158.63、189.34和148.55 mg·L-1。设计可扩增出猪、牛和熊的物种特异性引物。测序结果与GenBank数据库已登记序列同源性达99%以上。PCR体系各物种引物特异性强,无非特异扩增。应用于三重PCR体系中的引物互不干扰,检测灵敏度高,3种靶标样品任意一种DNA浓度降至1 mg·L-1时均可成功检测。27份人工模拟熊胆粉不同物种及不同比例掺假样品的检测结果与预设混合情况相符,盲法随机抽样重复检测5次,结果稳定。 成功建立可通过一次实验同时鉴别猪、牛和熊胆粉的方法,具有简便、灵敏及高效的特点,可应用于熊胆粉及其常见动物源性掺伪的鉴别。
中图分类号:
- R282.5