吉林大学学报(医学版) ›› 2022, Vol. 48 ›› Issue (6): 1546-1554.doi: 10.13481/j.1671-587X.20220621
基于乳酸代谢基因的肺腺癌预后模型的建立和评价
侯俊杰1,2,米旭光2,李孝男2,李晓男2,杨影2,芦小单2,方艳秋2,金宁一1,3( )
)
- 1.延边大学医学院病理生理学教研室,吉林 延吉133002
2.吉林省人民医院肿瘤综合治疗科,吉林 长春 130021
3.军事医学科学院分子病毒学与免疫学实验室,吉林 长春 130122
Establishment and evaluation of prognostic model of lung adenocarcinoma based on lactate metabolism gene
Junjie HOU1,2,Xuguang MI2,Xiaonan LI2,Xiaonan LI2,Ying YANG2,Xiaodan LU2,Yanqiu FANG2,Ningyi JIN1,3( )
)
- 1.Department of Pathophysiology,School of Medical Sciences,Yanbian University,Yanji 133002,China
2.Department of Comprehensive Oncology,Jilin Province People’s Hospital,Changchun 130021,China
3.Laboratory of Molecular Virology and Immunology,Academy of Military Medical Science,Changchun 130122,China
摘要:
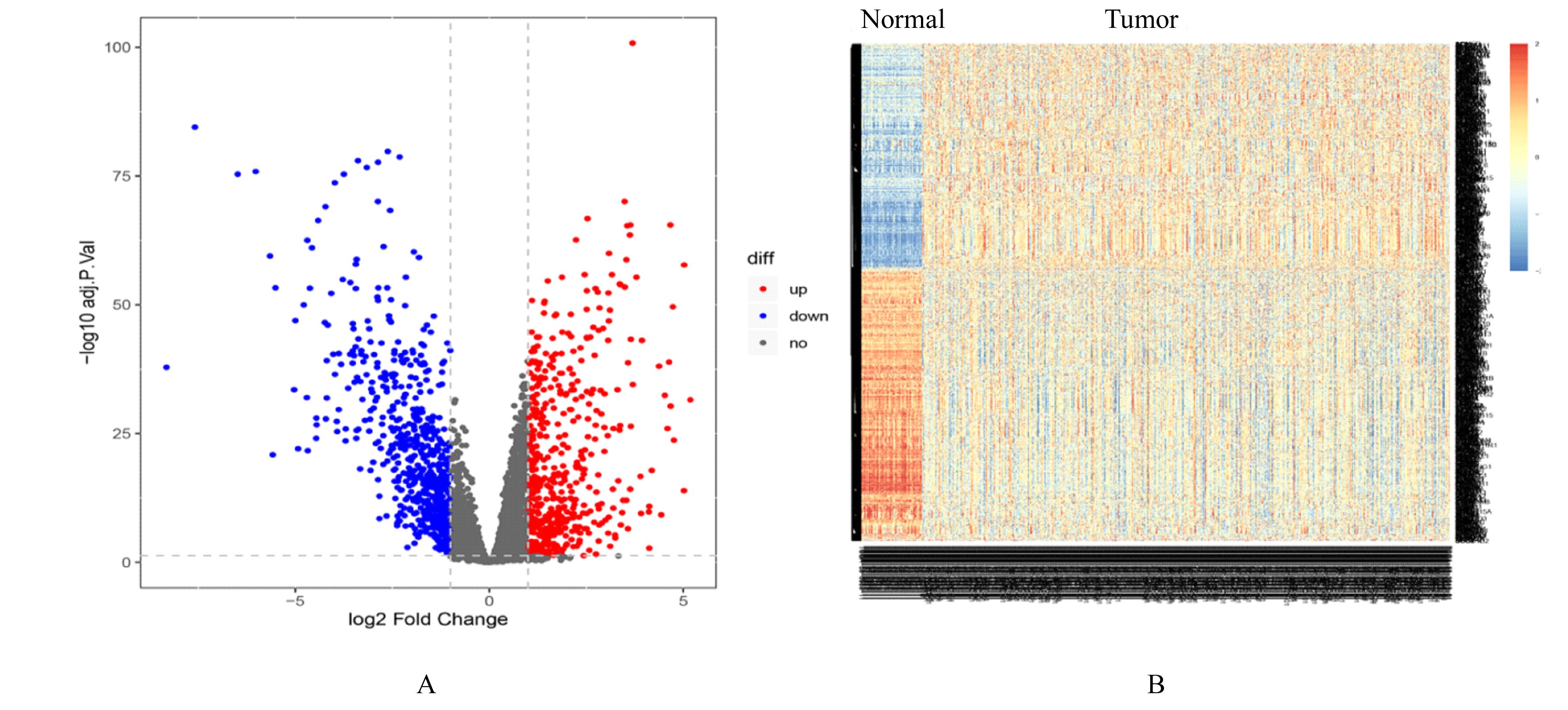
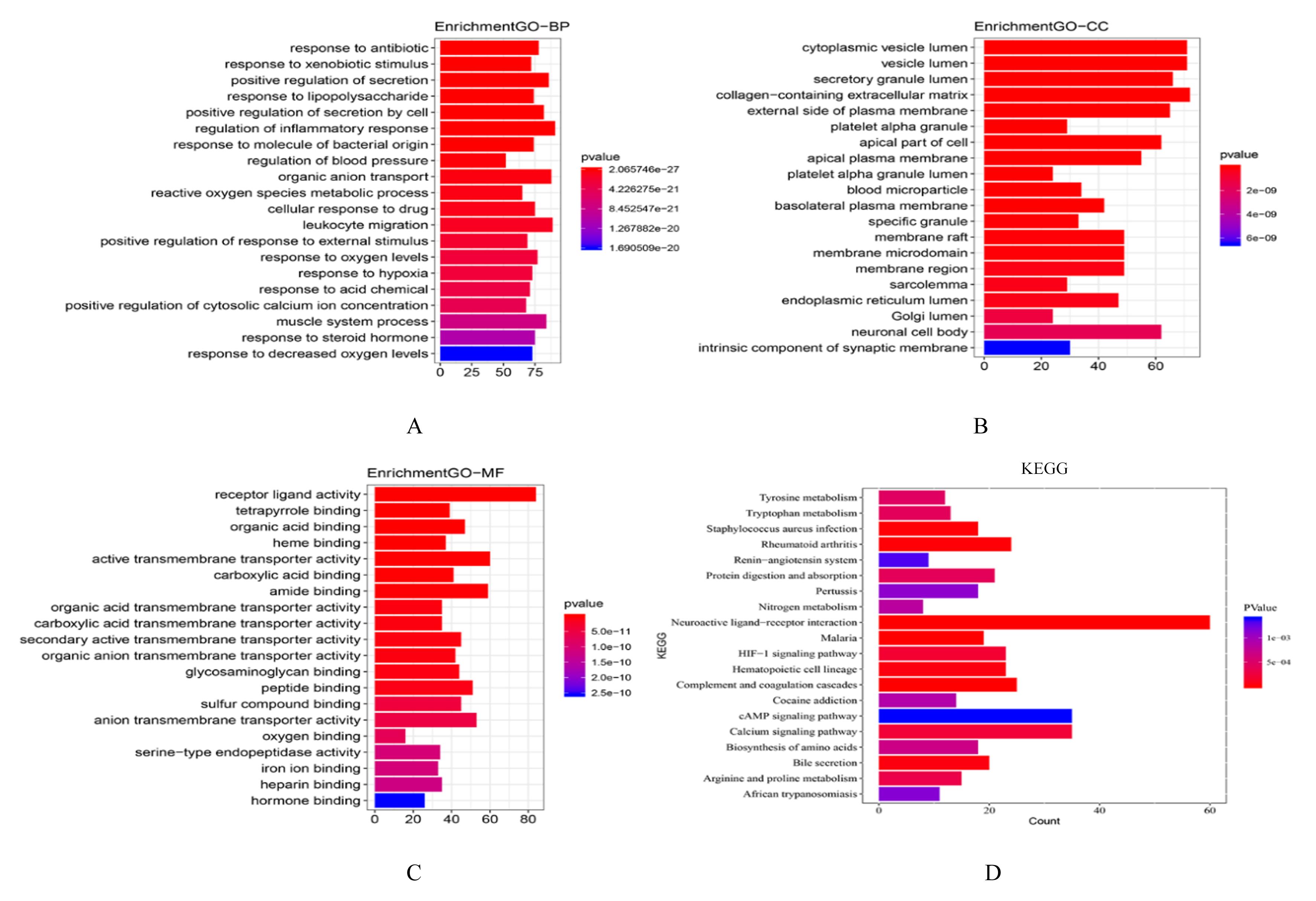
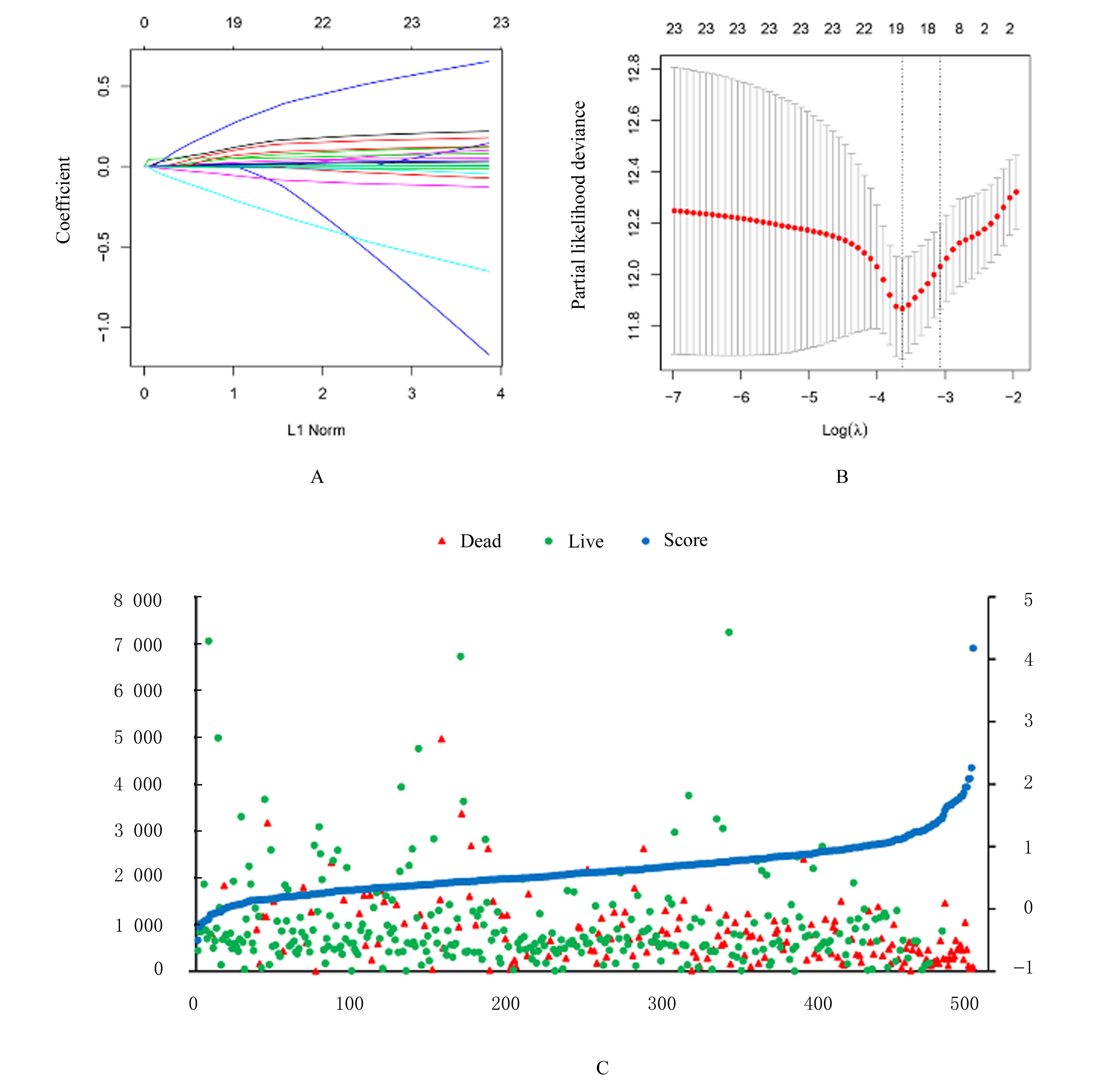
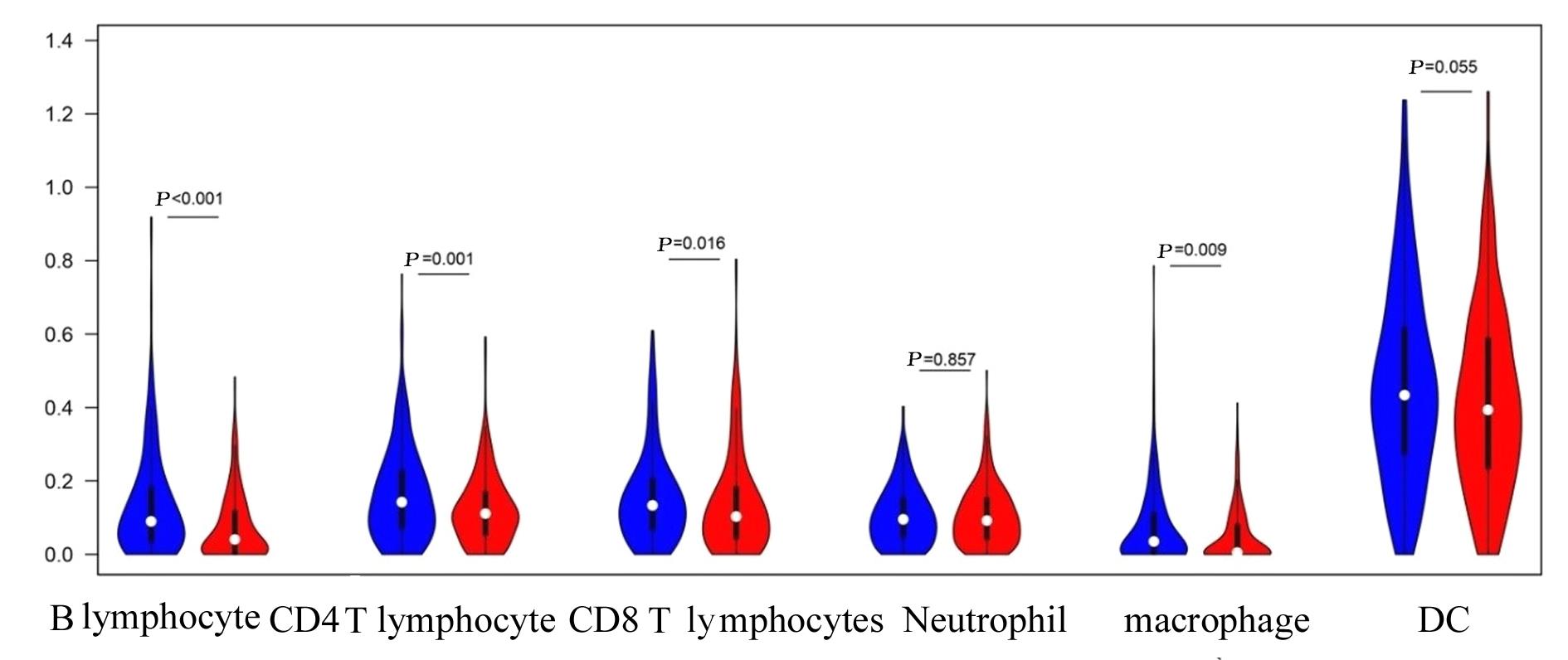
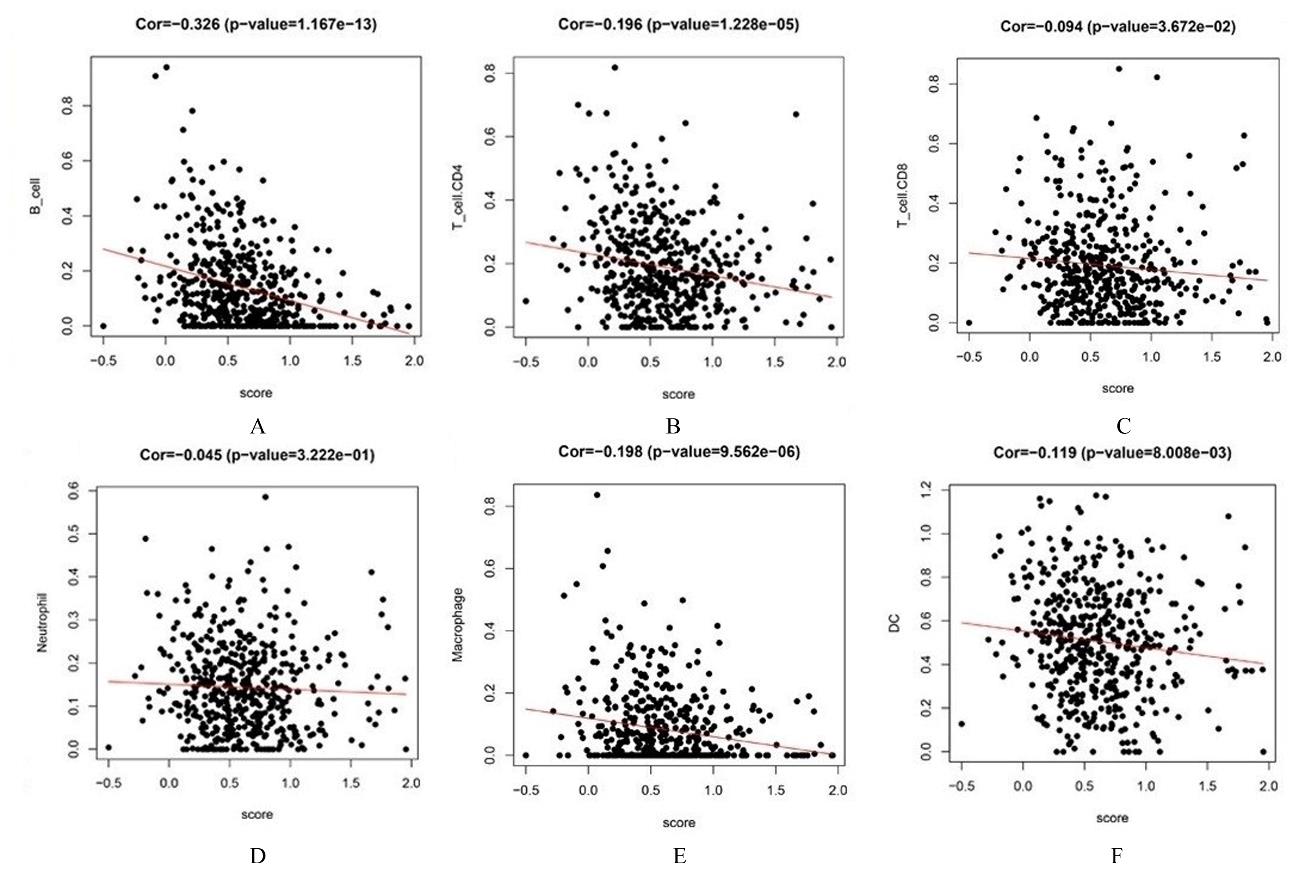
目的 探讨肺腺癌(LUAD)组织中乳酸代谢相关基因以及基于乳酸代谢基因构建LUAD预后评分模型,阐明其预测LUAD预后的能力。 方法 采用癌症基因图谱(TCGA)数据库筛选LUAD相关乳酸代谢基因。采用单因素和多因素Cox回归及LASSO回归分析获得关键基因并构建LUAD的乳酸代谢评分模型,采用Kaplan-Meier生存分析和受试者工作特征(ROC)曲线对模型的预测能力进行验证,TIMER法评估该模型与患者临床特征和免疫细胞浸润丰度之间的关系。 结果 成功筛选出16个乳酸代谢基因并构建评分模型;生存分析,低风险组患者的总生存时间(OS)明显高于高风险组(P< 0.01),且有较高的预测预后能力,ROC曲线下面积(AUC)均高于0.7;多因素Cox回归分析,23个乳酸代谢基因是LUAD患者的独立的预后基因;乳酸评分与B淋巴细胞(r=-0.326,P<0.001)、CD4 T淋巴细胞(r=-0.196,P<0.001)、CD8 T淋巴细胞(r=-0.094,P=0.036)、巨噬细胞(r=-0.198,P<0.001)和树突状细胞(r=-0.119,P=0.008)百分率呈负相关关系。 结论 乳酸代谢评分模型可以很好评估LUAD 预后与肿瘤微环境(TIMER)的状态,可作为LUAD预后预测的生物标志物。
中图分类号:
- R734.2