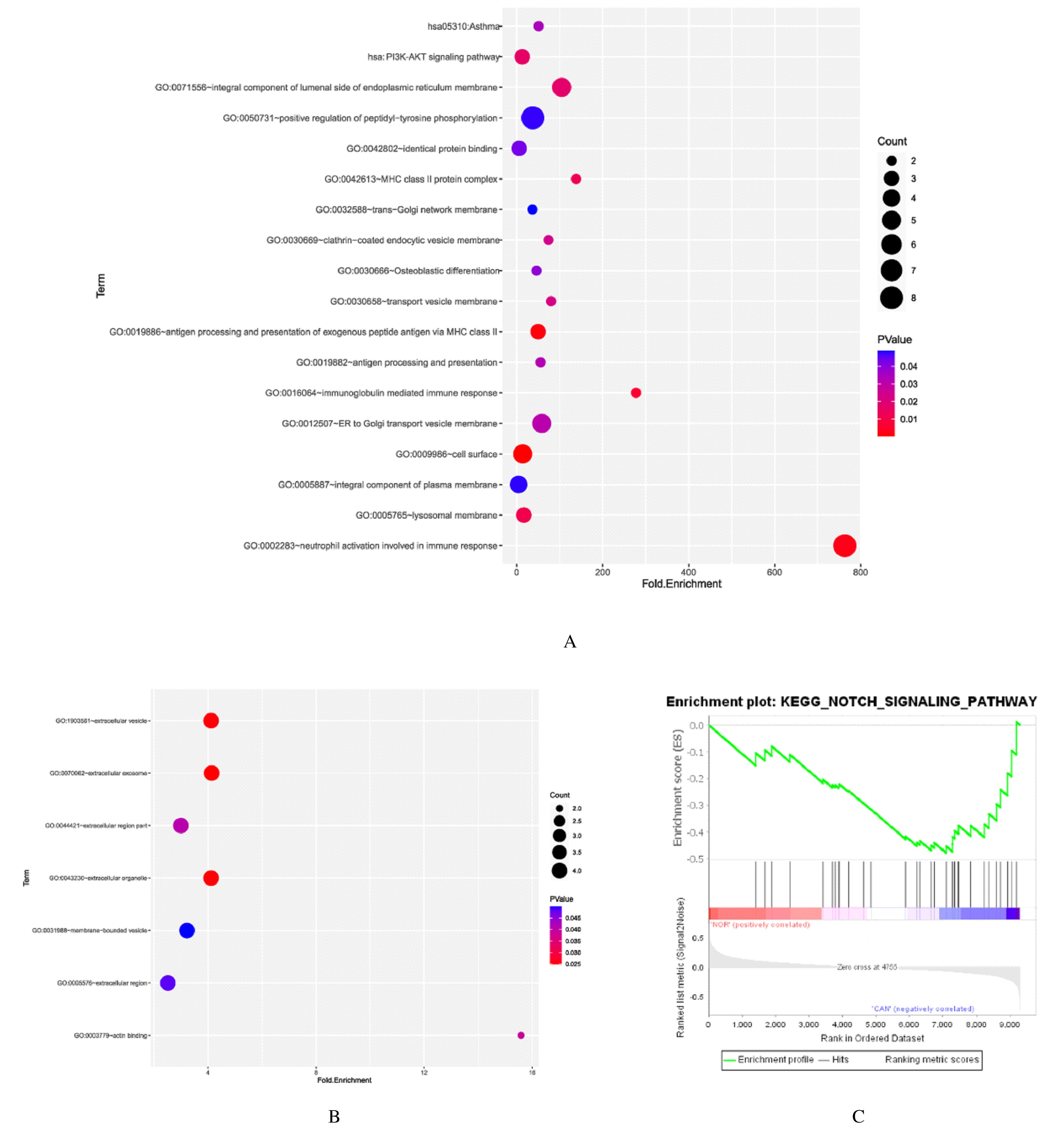
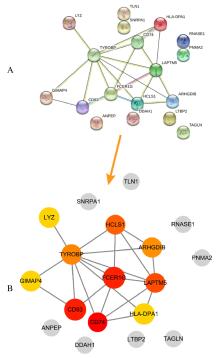
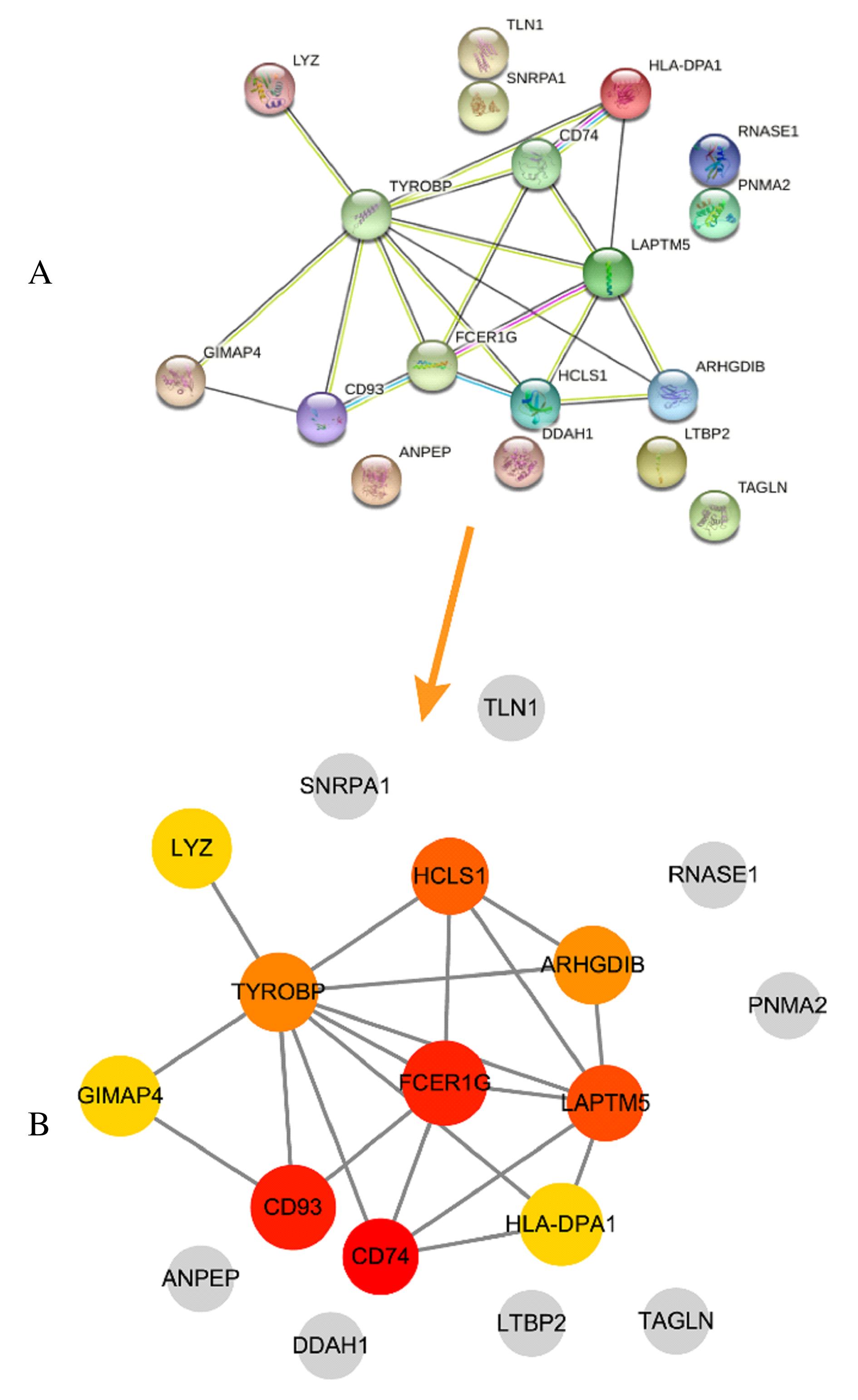
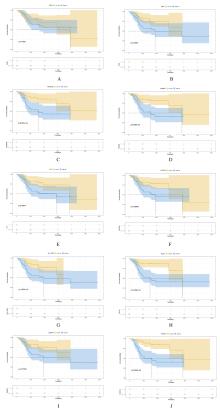
Journal of Jilin University(Medicine Edition) ›› 2021, Vol. 47 ›› Issue (6): 1570-1580.doi: 10.13481/j.1671-587X.20210631
• Methodology • Previous Articles Next Articles
Bioinformatics analysis based on screening of core driving genes in osteosarcoma and construction of gene model for prediction of survival time of patients
Weihang LI1,Ziyi DING1,Dong WANG1,Yikai PAN2,Yuhui LIU3,Shilei ZHANG1,Jing LI1,Ming YAN1( )
)
- 1.Department of Orthopaedics,Xijing Hospital,Air Force Medical University,Xi’an 710032,China
2.Department of Aerospace Medical Training,School of Aerospace Medicine,Air Force Medical University,Xi’an 710032,China
3.School of Aerospace Medicine,Center of Clinical Aerospace Medicine,Key Laboratory of Aerospace Medicine of Ministry of Education,Air Force Medical University,Xi’an 710032,China
-
Received:2021-04-20Online:2021-11-28Published:2021-12-14 -
Contact:Ming YAN E-mail:yanming_spine@163.com
CLC Number:
- R738.1
Cite this article
Weihang LI,Ziyi DING,Dong WANG,Yikai PAN,Yuhui LIU,Shilei ZHANG,Jing LI,Ming YAN. Bioinformatics analysis based on screening of core driving genes in osteosarcoma and construction of gene model for prediction of survival time of patients[J].Journal of Jilin University(Medicine Edition), 2021, 47(6): 1570-1580.
share this article
| 1 | GDOWSKI A S, RANJAN A, VISHWANATHA J K. Current concepts in bone metastasis, contemporary therapeutic strategies and ongoing clinical trials[J]. J Exp Clin Cancer Res, 2017, 36: 108. |
| 2 | MESSERSCHMITT P J, GARCIA R M, ABDUL-KARIM F W, et al. Osteosarcoma[J]. J Am Acad Orthop Surg, 2009, 17(8): 515-527. |
| 3 | LIN Y H, JEWELL B E, GINGOLD J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling[J]. Trends Mol Med, 2017, 23(8): 737-755. |
| 4 | LI W, YUAN B, ZHAO Y, et al. Transcriptome profiling reveals target in primary myelofibrosis together with structural biology study on novel natural inhibitors regarding JAK2[J]. Aging (Albany NY),2021,13(6): 8248-8275. |
| 5 | 何 远, 张丽娜, 朱兴文. 大数据技术在生物信息学中的应用综述[J]. 软件导刊, 2016, 15(4): 147-148. |
| 6 | GAUTIER L, COPE L, BOLSTAD B M, et al. Affy: analysis of Affymetrix GeneChip data at the probe level[J]. Bioinformatics, 2004, 20(3): 307-315. |
| 7 | COLAPRICO A, SILVA T C, OLSEN C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data[J]. Nucleic Acids Res, 2016, 44(8): e71. |
| 8 | DENNIS G, SHERMAN B T, HOSACK D A, et al. DAVID: database for annotation, visualization, and integrated discovery[J]. Genome Biol, 2003, 4(5): P3. |
| 9 | SZKLARCZYK D, GABLE A L, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res, 2019, 47(d1): D607-D613. |
| 10 | CHIN C H, CHEN S H, WU H H, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome[J]. BMC Syst Biol, 2014, 8(): S11. |
| 11 | ARNDT C A, ROSE P S, FOLPE A L, et al. Common musculoskeletal tumors of childhood and adolescence[J]. Mayo Clin Proc, 2012,87(5): 475-487. |
| 12 | SAMPO M, KOIVIKKO M, TASKINEN M, et al. Incidence, epidemiology and treatment results of osteosarcoma in Finland - a nationwide population-based study[J]. Acta Oncol, 2011, 50(8): 1206-1214. |
| 13 | PICCI P. Osteosarcoma (osteogenic sarcoma)[J]. Orphanet J Rare Dis, 2007, 2: 6. |
| 14 | XI L, ZHANG Y F, KONG S N, et al. miR-34 inhibits growth and promotes apoptosis of osteosarcoma in nude mice through targetly regulating TGIF2 expression[J]. Biosci Rep, 2018, 38(3):BSR20180078. |
| 15 | LUETKE A, MEYERS P A, LEWIS I, et al. Osteosarcoma treatment - Where do we stand? A state of the art review[J]. Cancer Treat Rev, 2014, 40(4): 523-532. |
| 16 | 任 娟, 徐叶峰, 匡唐洪, 等. 104例骨肉瘤肺转移患者的预后及其影响因素[J].中华肿瘤杂志, 2017,39(4): 263-268. |
| 17 | YANG L, LI W, ZHAO Y, et al. Computational study of novel natural inhibitors targeting O6-methylguanine-DNA methyltransferase[J]. World Neurosurg, 2019, 130: e294-e306. |
| 18 | ZHONG S, LI W H, BAI Y, et al. Computational study on new natural compound agonists of stimulator of interferon genes(STING)[J].PLoS One,2019,14(5):e0216678. |
| 19 | MOEK K L, DE GROOT D J A, DE VRIES E G E, et al. The antibody-drug conjugate target landscape across a broad range of tumour types[J]. Ann Oncol, 2017, 28(12): 3083-3091. |
| 20 | BUDDINGH E P, RUSLAN S E N, REIJNDERS C M A, et al. Mesenchymal stromal cells of osteosarcoma patients do not show evidence of neoplastic changes during long-term culture[J]. Clin Sarcoma Res, 2015, 5: 16. |
| 21 | PAN Y, LU L, CHEN J,et al. Identification of potential crucial genes and construction of microRNA-mRNA negative regulatory networks in osteosarcoma[J]. Hereditas, 2018, 155: 21. |
| 22 | HENCKAERTS L, CLEYNEN I, BRINAR M, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohnʼs disease[J]. Inflamm Bowel Dis, 2011, 17(6): 1392-1397. |
| 23 | BILALIC S, VEITINGER M, AHRER K H, et al. Identification of Non-HLA antigens targeted by alloreactive antibodies in patients undergoing chronic hemodialysis[J]. J Proteome Res, 2010, 9(2): 1041-1049. |
| 24 | LIU S S, CUI H, LI Q L, et al. RhoGDI2 is expressed in human trophoblasts and involved in their migration by inhibiting the activation of RAC1[J]. Biol Reprod, 2014, 90(4): 88. |
| 25 | ZHU H, WU H, LIU X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells[J]. Autophagy, 2009, 5(6): 816-823. |
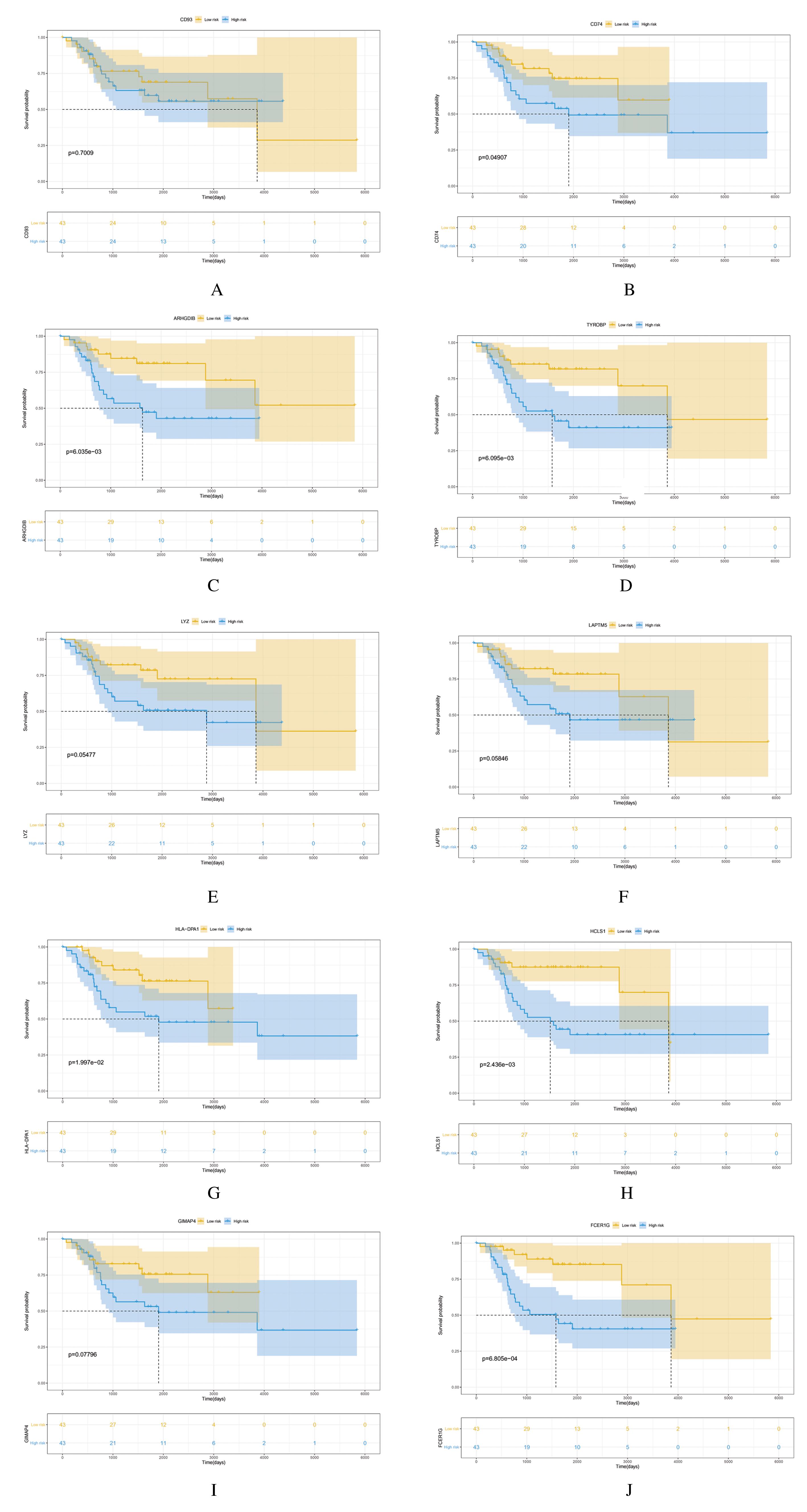
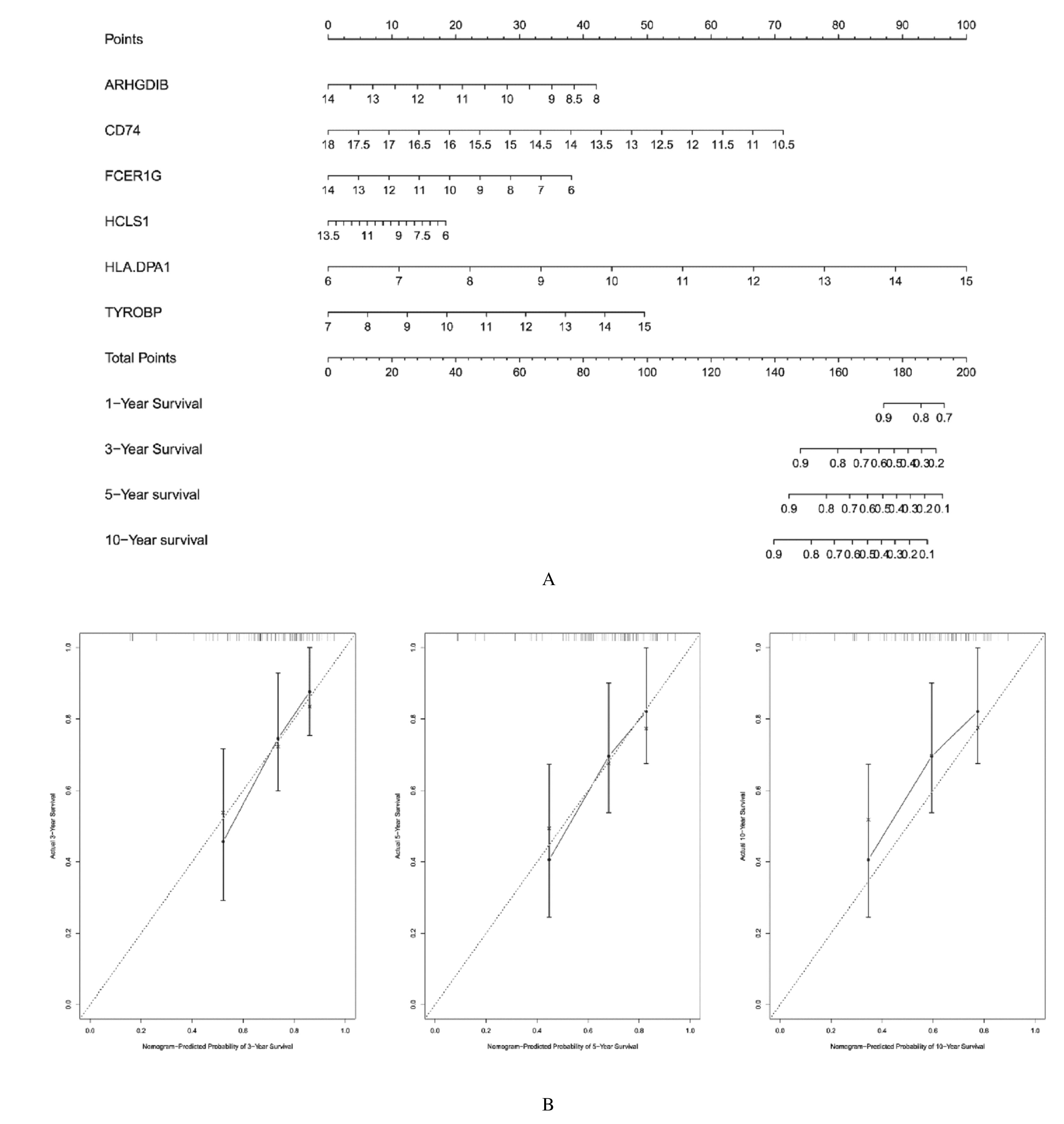
| 26 | SWEET R A, NICKERSON K M, CULLEN J L,et al. B cell–extrinsic Myd88 and Fcer1g negatively regulate autoreactive and normal B cell immune responses[J]. J Immunol, 2017, 199(3): 885-893. |
| 27 | LE CONIAT M, KINET J P, BERGER R. The human genes for the alpha and gamma subunits of the mast cell receptor for immunoglobulin E are located on human chromosome band 1q23[J]. Immunogenetics, 1990, 32(3): 183-186. |
| 28 | RAJARAMAN P, BRENNER A V, NETA G, et al. Risk of meningioma and common variation in genes related to innate immunity[J]. Cancer Epidemiol Biomarkers Prev, 2010, 19(5): 1356-1361. |
| 29 | MAHACHIE JOHN J M, BAURECHT H, RODRÍGUEZ E, et al. Analysis of the high affinity IgE receptor genes reveals epistatic effects of FCER1A variants on eczema risk[J]. Allergy, 2010, 65(7): 875-882. |
| 30 | HAN S, LAN Q, PARK A K, et al. Polymorphisms in innate immunity genes and risk of childhood leukemia[J]. Hum Immunol, 2010, 71(7): 727-730. |
| 31 | CHEN L, YUAN L, WANG Y, et al. Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma[J]. Int J Biol Sci, 2017, 13(11): 1361-1372. |
| 32 | PALONEVA J, KESTILÄ M, WU J, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts[J]. Nat Genet, 2000, 25(3): 357-361. |
| 33 | HAMERMAN J A, NI M J, KILLEBREW J R, et al. The expanding roles of ITAM adapters FcRgamma and DAP12 in myeloid cells[J]. Immunol Rev, 2009, 232(1): 42-58. |
| 34 | 施锦涛,张 凯,张芮浩,等.PD-1/PD-L1轴与骨肉瘤治疗研究进展[J].解放军医学杂志,2021,46(1):89-94. |
| 35 | HAURE-MIRANDE J V, AUDRAIN M, FANUTZA T, et al. Deficiency of TYROBP, an adapter protein for TREM2 and CR3 receptors, is neuroprotective in a mouse model of early Alzheimer’s pathology[J]. Acta Neuropathol, 2017, 134(5): 769-788. |
| [1] | Ping HU,Nan JIANG,Yinghua LI,Hongguang ZHAO. Analysis on characteristics of 18F-FDG PET/CT imaging of patient with primary small cell osteosarcoma of coccyx:A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 216-221. |
| [2] | CHEN Bingpeng, REN Ming, LI Ronghang, HAN Qing, WANG Jincheng. Regulatory effect of NOB1 on osteosarcoma cell-related genes detected by gene microarray technique [J]. Journal of Jilin University(Medicine Edition), 2018, 44(05): 929-934. |
| [3] | NA Jian, DAI Weixiang, MA Chao, ZHANG Xiaodong, SHAO Changqing, LIU Yong, WANG Xiuli, LIU Ying. Inhibitory effect of fluorouracil combined with DDP on human osteosarcoma cell line MG-63 and its influence in expressions of TRPV5 and TRPV6 proteins [J]. Journal of Jilin University Medicine Edition, 2015, 41(06): 1201-1206. |
| [4] | CAI Wen-tao,XIA Hong,SHENG Ning-jiang,LIN Ming-xia. Inhibitory effect of transgiutaminase 2 on apoptosis of osteosarcoma cell line MG-63 and its mechanism [J]. Journal of Jilin University Medicine Edition, 2014, 40(04): 782-789. |
| [5] | ZHANG Zhi-yu,HU Shuo,CAI Zheng-dong. Photodynamic effects of BCPD-17 and PSD007 on osteosarcoma LM-8 transplanted in mice [J]. J4, 2012, 38(4): 644-648. |
| [6] | WAN Fa-qing, WANG Jing, SHAN Yu-xing, ZHANG Hai-yu. Inhibitory effect of telomerase inhibitor on proliferation of osteosarcoma cell line in vitro [J]. J4, 2012, 38(2): 254-258. |
| [7] | QIAN Ming,LIU Min,DUAN Meng-na,LIU Chang,HE Xi,ZHOU Yan-min. Influence of oxytocin in proliferation and osteogenesis |activity of MG-63 cells [J]. J4, 2012, 38(2): 262-265. |
| [8] |
WANG Jing, LIU Yi-Jun, ZHAO Ding, SHAN Yu-Xin.
Expressions of MMP-9 and PCNA in telomerase and alternative lengthening of telomere osteosarcoma and their clinical significances [J]. J4, 2011, 37(3): 495-498. |
| [9] | GE Yan, WANG Yi-Shu, GAO Ting, ZHANG Li-Hong, LIAO Xiang-Yu, HAO Yan-Yan, LI Yu-Lin. Inhibitory effect of mistletoe alkali on osteosarcoma cells [J]. J4, 2009, 35(5): 841-844. |
| [10] | LEI Na-Wei, BO Xiu-Jie, JI E-Ling, FANG Yan-Qiu. Expressions of P16 and proliferating cell nuclear antigen in osteosarcoma and clinical significances [J]. J4, 2009, 35(3): 519-521. |
| [11] | LU Jia-yin,WU Dan-kai,XU Chun-hua,ZHANG Duo-duo,WU Dan-hua,GAO Zhong-li,ZHAO Yan-ying. Inhibitory effects of siRNA on MDM2 expression and cell proliferation in osteosarcoma cell line [J]. J4, 2008, 34(3): 442-445. |
| [12] | YANG Xiao-yu,GU Rui,GAO Zhong-li,SUI Fu-ge,SUN Wei,YIN Zhao-yang,ZHANG Shan-yong . Application and establishment of two-dimensional gel electrophoresis from proteome analysis of human osteosarcoma [J]. J4, 2007, 33(6): 1038-1042. |
| [13] | LU Jia-yin,WU Dan-kai,GAO Zhong-li,ZHAO Yan-ying,LIU Hui. Expressions of MDM2 and VEGF in osteosarcoma tissue and clinical significances [J]. J4, 2007, 33(3): 577-580. |
| [14] | QIN Da-ming,SUN Da-hui,GU Gui-shan,TAN Yan. Effects of semiconductor laser on proliferation of multidrug-resistantmodel of human osteosarcoma cell line (R-OS-732) [J]. J4, 2006, 32(2): 268-271. |
| [15] | LI Hong-wei, WANG Jin-cheng,BU Li-sha, DUAN De-sheng, SHI Jian-sheng. Preparation of monoclonal antibodies against humanosteogenic sarcoma and its characteristics [J]. J4, 2004, 30(4): 559-561. |