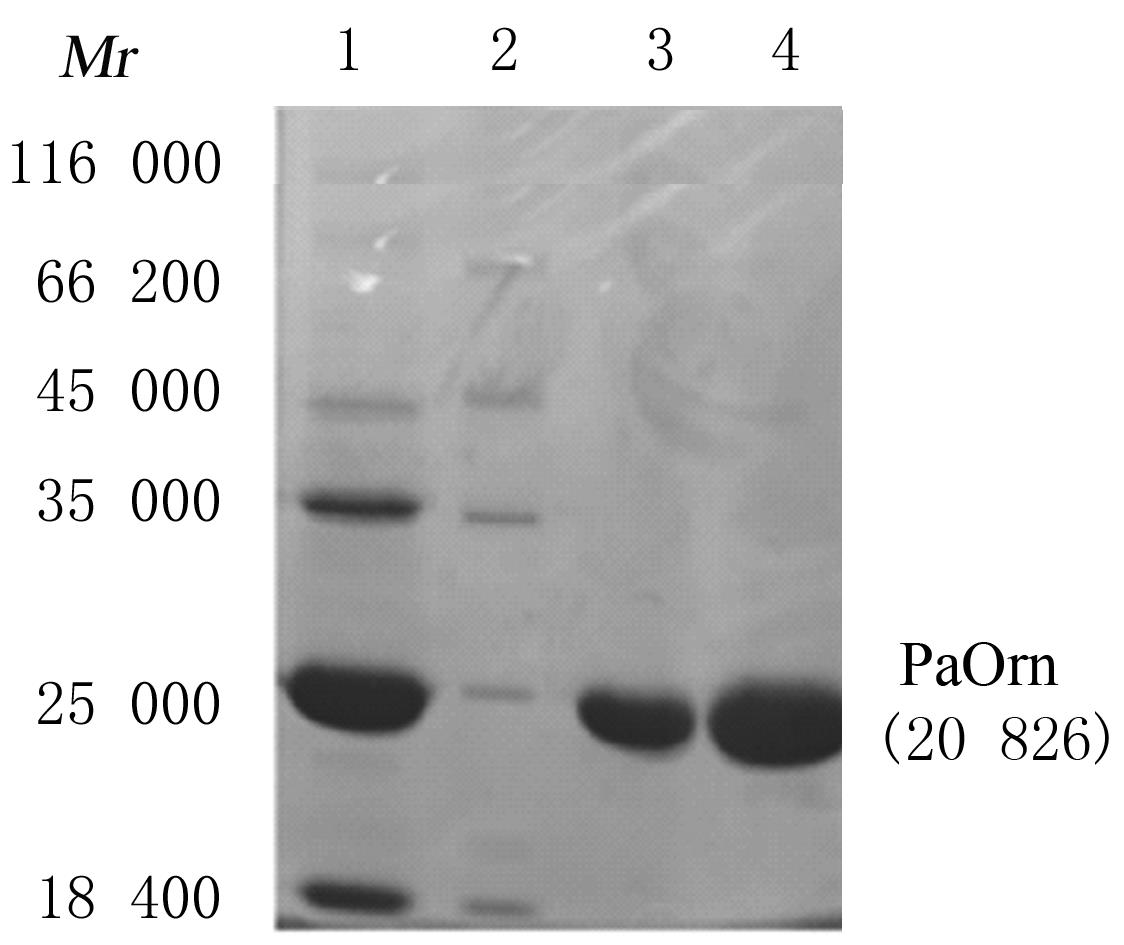
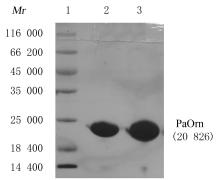
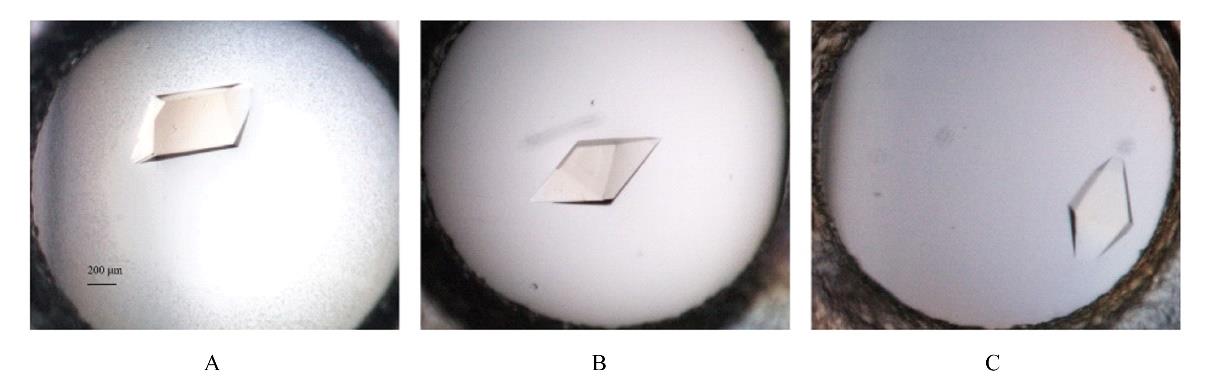
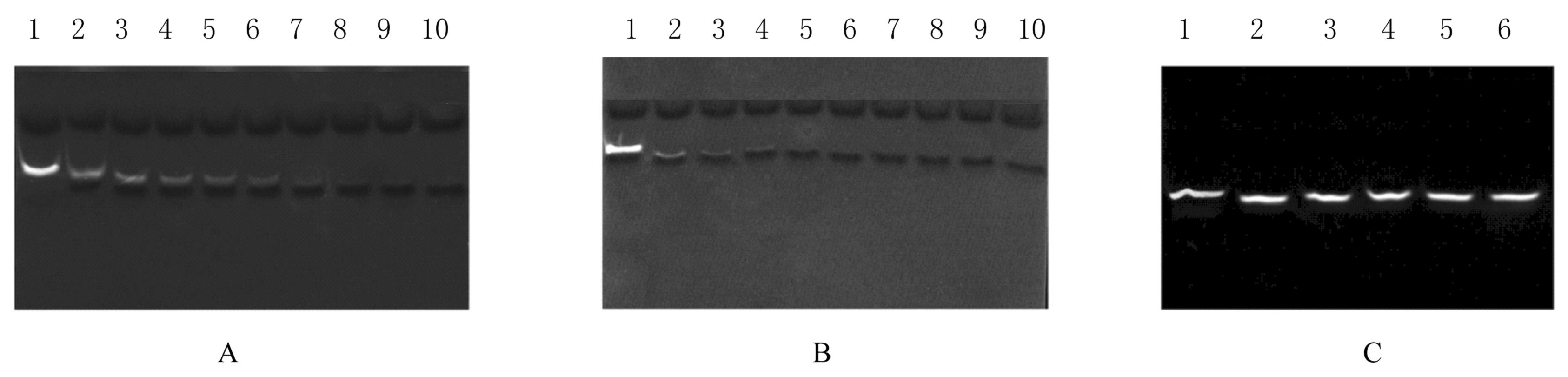
| 1 |
BERUBE B J, RANGEL S M, HAUSER A R. Pseudomonas aeruginosa: breaking down barriers[J]. Curr Genet, 2016, 62(1): 109-113.
|
| 2 |
KERR K G, SNELLING A M. Pseudomonas aeruginosa: a formidable and ever-present adversary[J]. J Hosp Infect, 2009, 73(4): 338-344.
|
| 3 |
KIELHOFNER M, ATMAR R L, HAMILL R J,et al. Life-threatening Pseudomonas aeruginosa infections in patients with human immunodeficiency virus infection[J]. Clin Infect Dis, 1992, 14(2): 403-411.
|
| 4 |
SILBY M W, WINSTANLEY C, GODFREY S A C, et al. Pseudomonas genomes: diverse and adaptable[J]. FEMS Microbiol Rev, 2011, 35(4): 652-680.
|
| 5 |
TACCONELLI E, CARRARA E, SAVOLDI A,et al. Discovery,research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis[J].Lancet Infect Dis,2018,18(3):318-327.
|
| 6 |
RÖMLING U, GALPERIN M Y, GOMELSKY M. Cyclic di-GMP:the first 25 years of a universal bacterial second messenger[J].Microbiol Mol Biol Rev,2013, 77(1):1-52.
|
| 7 |
ZHANG H N, XU Z W, JIANG H W, et al. Cyclic di-GMP regulates Mycobacterium tuberculosis resistance to ethionamide[J]. Sci Rep, 2017, 7(1): 5860.
|
| 8 |
ORR M W, DONALDSON G P, SEVERIN G B,et al. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover[J]. Proc Natl Acad Sci U S A, 2015, 112(36): E5048-E5057.
|
| 9 |
COHEN D, MECHOLD U, NEVENZAL H, et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa[J]. Proc Natl Acad Sci U S A, 2015,112(36):11359-11364.
|
| 10 |
MOSCOSO J A, MIKKELSEN H, HEEB S, et al. The Pseudomonas aeruginosa sensor RetS switches type Ⅲ and type Ⅵ secretion via c-di-GMP signalling[J]. Environ Microbiol, 2011, 13(12): 3128-3138.
|
| 11 |
TISCHLER A D, CAMILLI A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression[J]. Infect Immun, 2005, 73(9): 5873-5882.
|
| 12 |
MAKOWSKI L, BARDHAN J, GORE D, et al. Multi-wavelength anomalous diffraction using medium-angle X-ray solution scattering (MADMAX)[J]. Biophys J, 2012, 102(4): 927-933.
|
| 13 |
Summer H, Grämer R, Dröge P. Denaturing urea polyacrylamide gel electrophoresis (urea PAGE)[J]. J Vis Exp, 2009,32: 1485.
|
| 14 |
CRISAFULI F A P, RAMOS E B, ROCHA M S. Characterizing the interaction between DNA and GelRed fluorescent stain[J]. Eur Biophys J, 2015, 44(1/2): 1-7.
|
| 15 |
XIE C, ZHONG D F, YU K T, et al. Recent advances in metabolite identification and quantitative bioanalysis by LC-Q-TOF MS[J]. Bioanalysis,2012,4(8):937-959.
|
| 16 |
KIM S K, LORMAND J D, WEISS C A, et al. A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation[J]. eLife, 2019, 8: e46313.
|
| 17 |
LIAO H B, LIU M F, GUO X L. The special existences: nanoRNA and nanoRNase[J]. Microbiol Res, 2018, 207: 134-139.
|
| 18 |
SUTTON J M, ZAHAR N MEL, BARTLETT M G. Oligonucleotide anion adduct formation using negative ion electrospray ion-mobility mass spectrometry [J]. J Am Soc Mass Spectrom, 2021, 32(2): 497-508.
|
| 19 |
ORR M W, WEISS C A, SEVERIN G B, et al. A subset of exoribonucleases serve as degradative enzymes for pGpG in c-di-GMP signaling[J]. J Bacteriol, 2018, 200(24) : e00300-e00318.
|
| 20 |
CHU L, AGRAWAL S, CHEN Y P, et al. Structural insights into nanoRNA degradation by human Rexo2[J]. RNA, 2019, 25(6): 737-746.
|
| 21 |
MCCARTHY R R, VALENTINI M, FILLOUX A. Contribution of cyclic di-GMP in the control of type Ⅲ and type Ⅵ secretion in Pseudomonas aeruginosa [J]. Methods Mol Biol, 2017, 1657: 213-224.
|