Journal of Jilin University(Medicine Edition) ›› 2023, Vol. 49 ›› Issue (2): 298-307.doi: 10.13481/j.1671-587X.20230205
• Research in basic medicine • Previous Articles Next Articles
Regulatory effect of lncRNA SNHG17 on biological behavior of non-small cell lung cancer cells by targeting AEG1 through miR-384
Yunpeng LIU1,Zihao LIU1,Boming KANG2,Zhiguang YANG1( )
)
- 1.Department of Thoracic Surgery, First Hospital, Jilin University, Changchun 130021, China
2.Department of Pharmacy, School of Pharmacy, Jilin University, Changchun 130021, China
-
Received:2022-06-27Online:2023-03-28Published:2023-04-24 -
Contact:Zhiguang YANG E-mail:zgyang@jlu.edu.cn
CLC Number:
- R734.2
Cite this article
Yunpeng LIU,Zihao LIU,Boming KANG,Zhiguang YANG. Regulatory effect of lncRNA SNHG17 on biological behavior of non-small cell lung cancer cells by targeting AEG1 through miR-384[J].Journal of Jilin University(Medicine Edition), 2023, 49(2): 298-307.
share this article
Tab. 1
Expression levels of lncRNA SNHG17, cell viabilities and numbers of migration and invasion cells in various groups after silencing lncRNA SNHG17 or over-expression of lncRNA SNHG17"
| Group | lncRNA SNHG17 | Cell viability (η/%) | Number of migration cells | Number of invasion cells |
|---|---|---|---|---|
| si-NC (A549 cells) | 1.03±0.24 | 95.32±5.45 | 384.00±12.00 | 191.00±5.03 |
| si-SNHG17 (A549 cells) | 0.21±0.02* | 45.68±10.22* | 289.00±6.08* | 111.00±3.06* |
| pcDNA3.1-NC (H1299 cells) | 1.02±0.19 | 96.56±3.89 | 499.00±13.53 | 147.00±6.11 |
| pcDNA3.1-SNHG17 (H1299 cells) | 6.35±1.42△ | 136.23±10.93△ | 716.00±11.27△ | 271.00±7.21△ |
Tab. 2
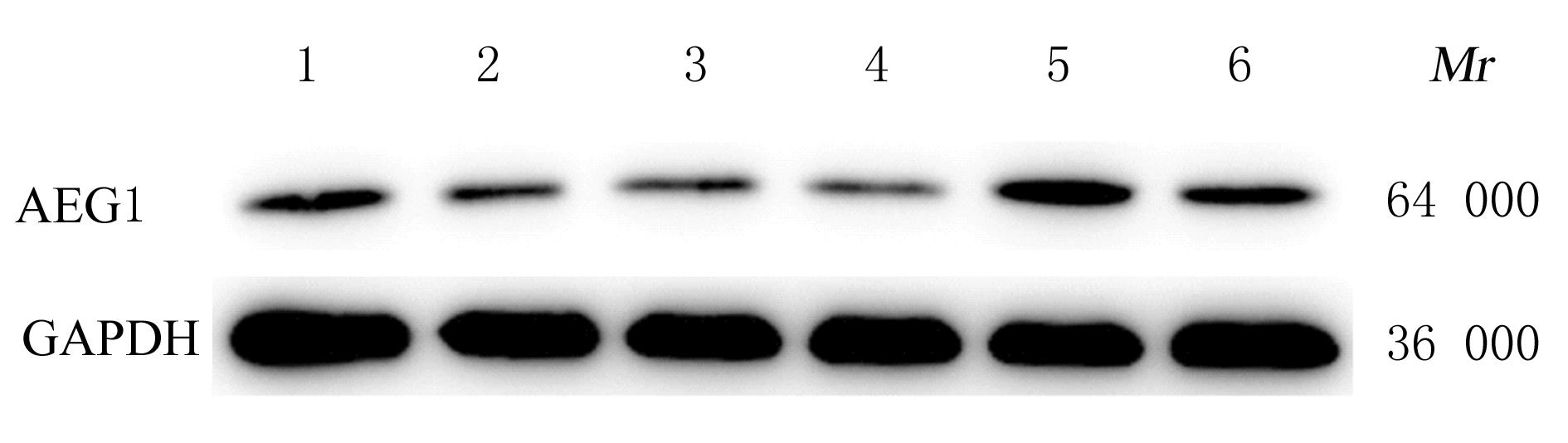
Expression levels of miR-384 and AEG1 mRNA in cells in various groups after silencing lncRNA SNHG17 or over-expression of lncRNA SNHG17"
| Group | miR-384 | AEG1 mRNA |
|---|---|---|
| si-NC(A549 cells) | 1.01±0.10 | 1.03±0.24 |
| si-SNHG17(A549 cells) | 1.46±0.20* | 0.06±0.02* |
| pcDNA3.1-NC(H1299 cells) | 1.01±0.13 | 1.02±0.19 |
| pcDNA3.1-SNHG17(H1299 cells) | 0.64±0.14△ | 2.34±0.27△ |
Tab. 3
Expression levels of miR-384, cell viabilities, numbers of migration and invasion cells in various groups"
| Group | miR-384 | Cell viability (η/%) | Number of migration cells | Number of invasion cells |
|---|---|---|---|---|
| Inhibitor NC(A549 cells) | 1.02±0.21 | 97.16±4.22 | 417.00±11.14 | 160.00±7.00 |
| miR-384 inhibitor(A549 cells) | 0.02±0.01* | 145.69±22.24* | 489.00±16.64* | 243.00±3.79* |
| Mimics NC(H1299 cells) | 1.02±0.21 | 96.37±3.22 | 518.00±11.53 | 207.00±6.03 |
| miR-384 mimics(H1299 cells) | 4.68±1.00△ | 65.67±6.40△ | 406.00±17.67△ | 116.00±8.00△ |
Tab. 4
Cell viabilities and numbers of migration and invasion cells in various groups after inhibiting or enhancing lncRNA SNHG17 and miR-384"
| Group | Cell viability (η/%) | Number of migration cells | Number ofinvasion cells | Expression level of AEG1 protein |
|---|---|---|---|---|
| si-NC+inhibitor NC(A549 cells) | 98.07±4.84 | 364.00±18.15 | 195.00±9.85 | 1.00±0.03 |
| si-SNHG17+inhibitor NC(A549 cells) | 63.26±6.04* | 314.00±8.72* | 108.00±6.11* | 0.72±0.01* |
| si-SNHG17+miR-384 inhibitor(A549 cells) | 78.69±6.65△ | 350.00±8.74△ | 199.00±11.15△ | 0.81±0.01△ |
| pcDNA3.1-NC+mimics NC(H1299 cells) | 101.26±5.91 | 496.00±18.23 | 148.00±4.58 | 1.00±0.02 |
| pcDNA3.1-SNHG17+mimics NC(H1299 cells) | 138.61±9.05# | 674.00±6.93# | 271.00±9.07# | 2.26±0.04# |
| pcDNA3.1-SNHG17+miR-384 mimics(H1299 cells) | 108.64±7.87○ | 582.00±14.53○ | 221.00±3.79○ | 1.56±0.02○ |
Tab. 5
Expression levels of AEG1 mRNA in cells, cell viabilities and numbers of migration and invasion cells in various groups after inhibiting or enhancing AEG1"
| Group | AEG1 mRNA | Cell viability (η/%) | Number of migration cells | Number of invasion cells |
|---|---|---|---|---|
| si-NC(A549 cells) | 1.02±0.22 | 98.67±4.76 | 381.00±13.08 | 167.00±6.03 |
| si-AEG1(A549 cells) | 0.56±0.08* | 71.32±8.98* | 308.00±10.26* | 115.00±10.02* |
| pcDNA3.1-NC(H1299 cells) | 1.01±0.11 | 99.21±7.66 | 499.00±11.53 | 137.00±7.09 |
| pcDNA3.1-AEG1(H1299 cells) | 3.15±0.90△ | 132.32±9.33△ | 609.00±11.14△ | 282.00±3.79△ |
Tab. 6
Cell viabilities and numbers of migration and invasion cells in various groups after inhibiting or enhancing miR-384 and AEG1 mRNA"
| Group | Cell viability (η/%) | Number of migration cells | Number of invasion cells |
|---|---|---|---|
| Inhibitor NC+si-NC(A549 cells) | 101.36±7.40 | 408.00±12.58 | 161.00±6.11 |
| miR-384 inhibitor+si-NC(A549 cells) | 145.37±19.97* | 481.00±8.66* | 256.00±9.17* |
| miR-384 inhibitor+si-AEG1(A549 cells) | 112.36±7.47△ | 425.00±9.07△ | 172.00±9.07△ |
| Mimics NC+pcDNA3.1-NC(H1299 cells) | 98.63±8.83 | 520.00±8.14 | 162.00±6.03 |
| miR-384 mimics+pcDNA3.1-NC(H1299 cells) | 63.17±9.12# | 401.00±10.44# | 117.00±2.08# |
| miR-384 mimics+pcDNA3.1-AEG1(H1299 cells) | 81.06±5.75○ | 589.00±6.24○ | 142.00±8.71○ |
| 1 | BALATA H, FONG K M, HENDRIKS L E, et al. Prevention and early detection for NSCLC: advances in thoracic oncology 2018 [J].J Thorac Oncol,2019,14(9): 1513-1527. |
| 2 | SUN J, ZHANG Z C, BAO S Q, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer [J]. J Immunother Cancer, 2020, 8(1):e000110. |
| 3 | LI X Y, LI Y, YU X M, et al. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer [J]. J Transl Med, 2020, 18(1): 331. |
| 4 | LUO Y H, ZHENG S T, WU Q Y, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation [J]. Autophagy, 2021, 17(12): 4083-4101. |
| 5 | GHAFOURI-FARD S, SHOOREI H, BRANICKI W, et al. Non-coding RNA profile in lung cancer [J]. Exp Mol Pathol, 2020, 114: 104411. |
| 6 | WANG H B, MENG Q H, QIAN J J, et al. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer [J]. Pharmacol Ther, 2022, 234: 108123. |
| 7 | MA L, GAO J, ZHANG N, et al. Long noncoding RNA SNHG17: a novel molecule in human cancers [J]. Cancer Cell Int, 2022, 22(1): 104. |
| 8 | MA Z H, GU S Y, SONG M, et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer [J]. Mol Biosyst,2017,13(11): 2350-2361. |
| 9 | XU T W, YAN S, JIANG L H, et al. Gene Amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer [J]. Mol Ther Nucleic Acids, 2019, 17: 405-413. |
| 10 | MA T, ZHOU X J, WEI H L, et al. Long Non-coding RNA SNHG17 upregulates RFX1 by sponging miR-3180-3p and promotes cellular function in hepatocellular carcinoma [J]. Front Genet, 2020, 11: 607636. |
| 11 | ZHANG Y T, YANG G Y. LncRNA SNHG17 acts as a ceRNA of miR-324-3p to contribute the progression of osteosarcoma [J]. J Biol Regul Homeost Agents, 2020, 34(4): 1529-1533. |
| 12 | RELLI V, TREROTOLA M, GUERRA E, et al. Abandoning the notion of non-small cell lung cancer [J]. Trends Mol Med, 2019, 25(7): 585-594. |
| 13 | WANG M N, HERBST R S, BOSHOFF C. Toward personalized treatment approaches for non-small-cell lung cancer [J]. Nat Med, 2021, 27(8): 1345-1356. |
| 14 | WANG R Q, LONG X R, ZHOU N N, et al. Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging miR-26a-5p to activate PTEN signaling in non-small cell lung cancer [J]. J Exp Clin Cancer Res, 2021, 40(1): 9. |
| 15 | TONG F, GUO J, MIAO Z Q, et al. LncRNA SNHG17 promotes the progression of oral squamous cell carcinoma by modulating miR-375/PAX6 axis [J]. Cancer Biomark, 2021, 30(1): 1-12. |
| 16 | CHEN W H, WANG L F, LI X Y, et al. LncRNA SNHG17 regulates cell proliferation and invasion by targeting miR-338-3p/SOX4 axis in esophageal squamous cell carcinoma [J]. Cell Death Dis, 2021, 12(9): 806. |
| 17 | ZHANG Z W, YAN Y L, ZHANG B, et al. Long non-coding RNA SNHG17 promotes lung adenocarcinoma progression by targeting the microRNA-193a-5p/NETO2 axis [J]. Oncol Lett, 2021, 22(6): 818. |
| 18 | LI W, ZHENG Y Q, MAO B, et al. SNHG17 upregulates WLS expression to accelerate lung adenocarcinoma progression by sponging miR-485-5p[J]. Biochem Biophys Res Commun, 2020, 533(4): 1435-1441. |
| 19 | FAN N, ZHANG J, CHENG C T, et al. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling [J]. Biomed Pharmacother, 2017, 95: 1331-1337. |
| 20 | ZHONG B Z, WANG Q, LIU F, et al. Effects of miR-384 and miR-134-5p acting on YY1 signaling transduction on biological function of gastric cancer cells [J]. Onco Targets Ther, 2020, 13: 9631-9641. |
| 21 | TIAN Y H, JIA L W, LIU Z F, et al. LINC01087 inhibits glioma cell proliferation and migration, and increases cell apoptosis via miR-384/Bcl-2 axis [J]. Aging (Albany NY), 2021, 13(16): 20808-20819. |
| 22 | SUN H, CHEN Y, FANG Y Y, et al. Circ_0000376 enhances the proliferation, metastasis, and chemoresistance of NSCLC cells via repressing miR-384 [J]. Cancer Biomark, 2020, 29(4): 463-473. |
| 23 | GUO Q K, ZHENG M, XU Y, et al. MiR-384 induces apoptosis and autophagy of non-small cell lung cancer cells through the negative regulation of Collagen α-1(X) chain gene [J]. Biosci Rep, 2019,39(2):BSR20181523. |
| 24 | LI Y H, XU C L, HE C J, et al. circMTDH.4/miR-630/AEG-1 axis participates in the regulation of proliferation, migration, invasion, chemoresistance, and radioresistance of NSCLC [J]. Mol Carcinog,2020,59(2): 141-153. |
| 25 | SRIRAMULU S, SUN X F, MALAYAPERUMAL S, et al. Emerging role and clinicopathological significance of AEG-1 in different cancer types: a concise review [J]. Cells, 2021, 10(6):1497. |
| [1] | Xiaomei HUANG,Hongwei WANG,Zhenyan HAN,Qiuyuan WEI,Sujing HUANG. Effects of lncRNA GHET1 on biological behaviors of trophoblast cells in preeclampsia through Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1324-1332. |
| [2] | Zhihui ZHAO,Xianghua BAI,Jinling HE,Weiqin DUAN,Min LIU,Shengmao ZHANG. Inhibitory effect of sufentanil on apoptosis of myocardial cells in myocardial ischemia-reperfusion injury rats and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 364-373. |
| [3] | Junjie HOU,Xuguang MI,Xiaonan LI,Xiaonan LI,Ying YANG,Xianzhuo JIANG,Ying ZHOU,Zhiqiang NI,Ningyi JIN,Yanqiu FANG. Bronchopleural fistula complicated in treatment process of non-small cell lung cancer by bevacizumab combined with paclitaxel: A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 513-517. |
| [4] | Chaofeng ZHOU,Shifan ZHOU,Qing TIAN,Sai WANG,Honglin LI,Chunzheng MA. Effect of lncRNA-NORAD overexpression on biological behaviors of esophageal cancer Eca-109 cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 33-43. |
| [5] | Ye YUAN,Jinbao ZHUANG,Xu SHI,Changyuan LI. Influence of PGE2 on PD-1 expression in infiltrating T lymphocytes in non-small cell lung cancer tissue and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 249-256. |
| [6] | Guangcai WAN,Hongshuai SUN,Hua ZHU,Xiuyan YU. Meta-analysis on relationship between neutrophil/lymphocyte ratio and prognosis of patients with non-small cell lung cancer treated with EGFR-TKIs [J]. Journal of Jilin University(Medicine Edition), 2020, 46(6): 1267-1273. |
| [7] | QIAO Liangwei, QU Qingshan, LI Ming. Effect of LncRNA-ATB on acute rejection of kidney transplanted rats by regulating miR-200c expression [J]. Journal of Jilin University(Medicine Edition), 2020, 46(05): 955-962. |
| [8] | GUO Weiwei, QIN Yue, YANG Haibo, MI Zhanhu. Promotion effect of LncRNA MALAT1 on osteogenic differentiation of adipose-derived mesenchymal stem cells through miR-34c/SATB2 axis [J]. Journal of Jilin University(Medicine Edition), 2020, 46(05): 963-971. |
| [9] | WEI Xujing, LI Lin, ZHANG Hongzhen, WANG Jing, XU Jing. Effects of LncRNA CCAT1 on proliferation,invasion and migration of endometrial cancer cells through TGF-β1/smad signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2020, 46(05): 1016-1022. |
| [10] | DAI Juanjuan, YANG Lijuan, DU Jing, MIAO Shuang, XI Sichuan, LI Chen, WU Yan. Construction of PC9 cell lines stably transfectedwith human LncRNAMIR31HG gene and its effects on cell proliferation and migration [J]. Journal of Jilin University(Medicine Edition), 2019, 45(04): 801-806. |
| [11] | HUO Xiaolei, PEI Zhen, YANG Hao, ZHANG Yiqiang, JIA Jiantao, HAN Lingna. Effect of LncRNA-BLACAT1 on cell proliferation of non-small cell lung cancer through regulation of CyclinD1/CDKN2B axis and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2019, 45(04): 759-765. |
| [12] | WANG Zhijing, SU Rongjian, DU Xiaoyuan. Effects of GRP78 on sensibility of gemcitabine on patients with NSCLC [J]. Journal of Jilin University(Medicine Edition), 2019, 45(03): 595-600. |
| [13] | XU Jinrui, LYU Cuiping, YANG Yi, ZHOU Yanbing, LUO Jia, WANG Yujiong. Transcriptomesequencing and analysis of bovine alveolar macrophages infected with BCG [J]. Journal of Jilin University(Medicine Edition), 2019, 45(01): 12-16. |
| [14] | LI Lijing, ZANG Yaru, YU Wenhui, LIANG Junwei, TONG Yanjun. Therapeutic effect of chemotherapy combined with sodium cantharidinate vitamin B6 injection in patientswith non-small cell lung cancer and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2018, 44(06): 1286-1290. |
| [15] | YANG Wenyan, LIU Qiang, SUN Zhijuan, DU Liqing, XU Chang, WANG Yan, LIU Yang, WANG Qin. Effect of melatonin on radiosensitivity of non-small cell lung cancer H1299 cells [J]. Journal of Jilin University Medicine Edition, 2018, 44(03): 532-536. |
|
||