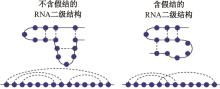
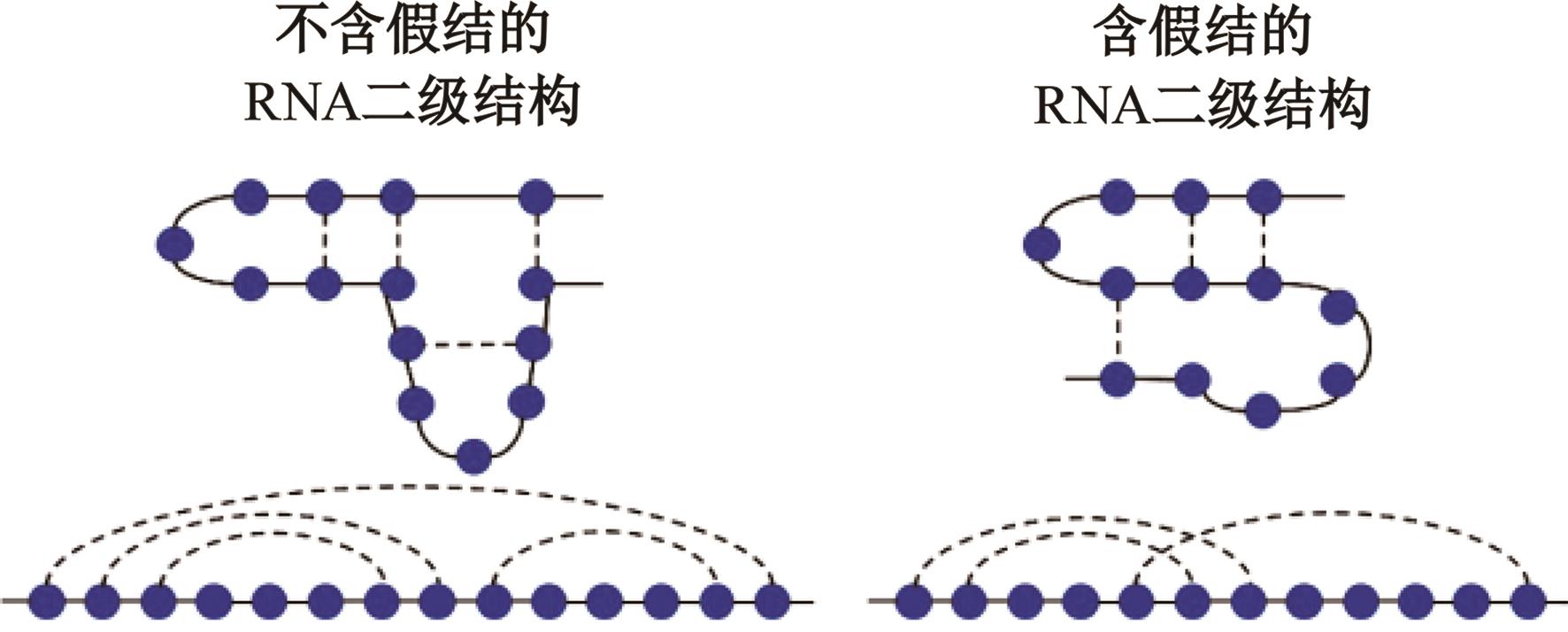
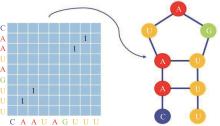
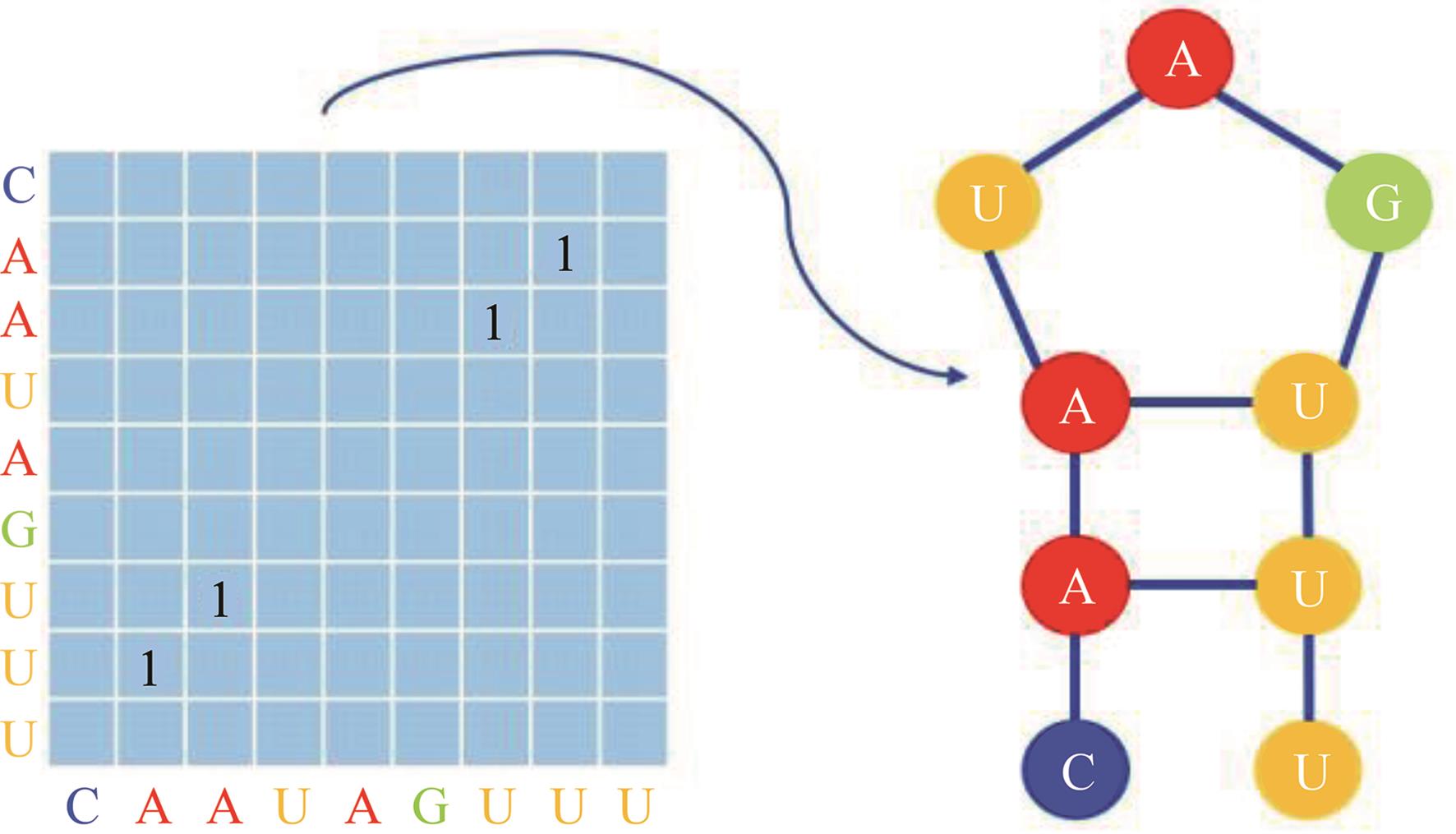
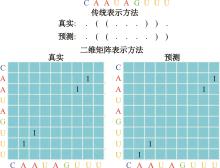
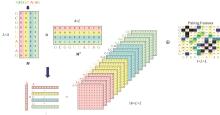
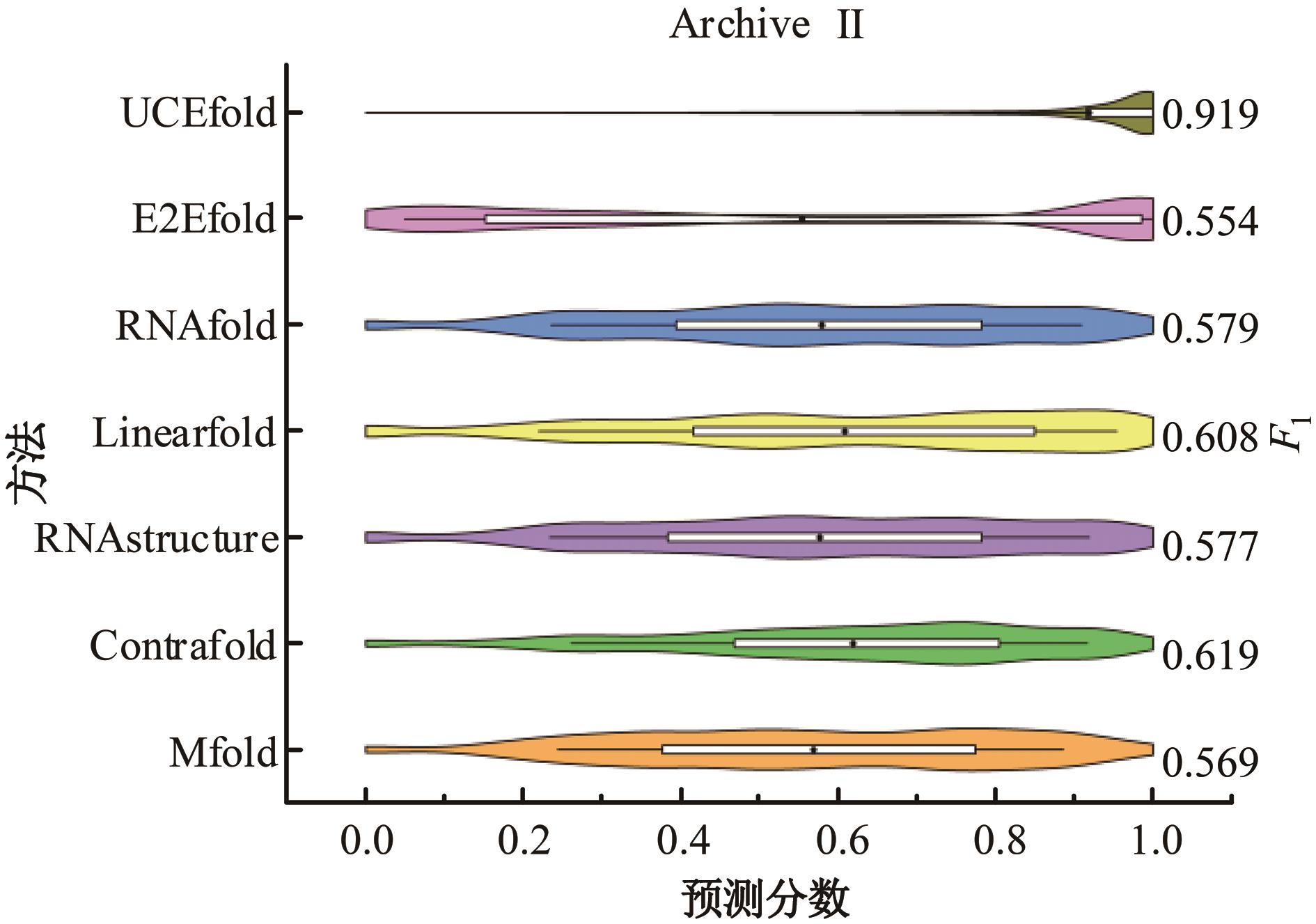
Journal of Jilin University(Engineering and Technology Edition) ›› 2025, Vol. 55 ›› Issue (1): 297-306.doi: 10.13229/j.cnki.jdxbgxb.20230267
Deep learning-based method for ribonucleic acid secondary structure prediction
Yuan-ning LIU1,2( ),Zi-nan ZANG1,2,Hao ZHANG1,2(
),Zi-nan ZANG1,2,Hao ZHANG1,2( ),Zhen LIU1,3
),Zhen LIU1,3
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education,Jilin University,Changchun 130012,China
3.Graduate School of Engineering,Nagasaki Institute of Applied Science,Nagasaki 851-0193,Japan
CLC Number:
- TP399
| 1 | Crick F. Central dogma of molecular biology[J]. Nature, 1970, 227: 561-563. |
| 2 | Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription[J]. Science, 2007, 316: 1484-1488. |
| 3 | Sharp P. The centrality of RNA[J]. Cell, 2009, 136: 577-580. |
| 4 | Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction[J]. Nucleic Acids Research, 2003, 31: 3406-3415. |
| 5 | Lorenz R, Bernhart S H, Höner Zu Siederdissen C, et al. ViennaRNA Package 2.0[J]. Algorithms for Molecular Biology, 2011, 6: No.26. |
| 6 | Mathews D H, Turner D H. Prediction of RNA secondary structure by free energy minimization[J]. Current Opinion in Structural Biology, 2006, 16: 270-278. |
| 7 | Huang L, Zhang H, Deng D, et al. LinearFold: linear-time approximate RNA folding by 5'-to-3' dynamic programming and beam search[J]. Bioinformatics, 2019, 35: i295-i304. |
| 8 | Brierley I, Pennell S, Gilbert R J C. Viral RNA pseudoknots: versatile motifs in gene expression and replication[J]. Nature Reviews Microbiology, 2007, 5: 598-610. |
| 9 | Bernhart S H, Hofacker I L, Will S, et al. RNAalifold: improved consensus structure prediction for RNA alignments[J]. BMC Bioinformatics, 2008, 9: No.474. |
| 10 | Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars[J]. Nucleic Acids Research, 2003, 31: 3423-3428. |
| 11 | Do C B, Woods D A, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models[J]. Bioinformatics, 2006, 22: e90-e98. |
| 12 | Zakov S, Goldberg Y, Elhadad M, et al. Rich parameterization improves RNA structure prediction. [J]. Journal of Computational Biology: A Journal of Computational Molecular Cell Biology, 2011, 6577: 546-562. |
| 13 | Zhang H, Zhang C, Li Z, et al. A new method of RNA secondary structure prediction based on convolutional neural network and dynamic programming[J]. Frontiers in Genetics, 2019, 10: No.467 |
| 14 | Chen X, Li Y, Umarov R, et al. RNA secondary structure prediction by learning unrolled algorithms [C]∥Proceedings of the International Conference on Learning Representations(ICLR), Addis Ababa, Ethiopia, 2020: 1-19. |
| 15 | Sato K, Akiyama M, Sakakibara Y. RNA secondary structure prediction using deep learning with thermodynamic integration[J]. Nature Communications, 2021, 12(1): No. 941. |
| 16 | Singh J, Hanson J, Paliwal K, et al. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning[J]. Nature Communications, 2019, 10(1): No. 5407. |
| 17 | Tan Z, Fu Y, Sharma G, et al. TurboFold Ⅱ: RNA structural alignment and secondary structure prediction informed by multiple homologs[J]. Nucleic Acids Research, 2017, 45(20): 11570-11581. |
| 18 | Sloma M, Mathews D. Exact calculation of loop formation probability identifies folding motifs in RNA secondary structures[J]. RNA, 2016, 22(12): 1808-1818. |
| 19 | Hochreiter S, Schmidhuber J. Long short-term memory[J]. Neural Computation, 1997, 9: 1735-1780. |
| 20 | Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need[C]∥Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, USA, 2017: 6000-6010. |
| 21 | Fu L, Cao Y, Wu J, et al. UFold: fast and accurate RNA secondary structure prediction with deep learning[J]. Nucleic Acids Research, 2022, 50(3): No.e14. |
| 22 | Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation [C]∥Proceedings of the Medical Image Computing and Computer-Assisted Intervention(MICCAI), Munich, Germany, 2015: 234-241. |
| [1] | Hui-zhi XU,Shi-sen JIANG,Xiu-qing WANG,Shuang CHEN. Vehicle target detection and ranging in vehicle image based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(1): 185-197. |
| [2] | Lu Li,Jun-qi Song,Ming Zhu,He-qun Tan,Yu-fan Zhou,Chao-qi Sun,Cheng-yu Zhou. Object extraction of yellow catfish based on RGHS image enhancement and improved YOLOv5 network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(9): 2638-2645. |
| [3] | Lei ZHANG,Jing JIAO,Bo-xin LI,Yan-jie ZHOU. Large capacity semi structured data extraction algorithm combining machine learning and deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(9): 2631-2637. |
| [4] | Hong-wei ZHAO,Hong WU,Ke MA,Hai LI. Image classification framework based on knowledge distillation [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(8): 2307-2312. |
| [5] | Yun-zuo ZHANG,Yu-xin ZHENG,Cun-yu WU,Tian ZHANG. Accurate lane detection of complex environment based on double feature extraction network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(7): 1894-1902. |
| [6] | Bai-you QIAO,Tong WU,Lu YANG,You-wen JIANG. A text sentiment analysis method based on BiGRU and capsule network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(7): 2026-2037. |
| [7] | Xin-gang GUO,Ying-chen HE,Chao CHENG. Noise-resistant multistep image super resolution network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(7): 2063-2071. |
| [8] | Li-ping ZHANG,Bin-yu LIU,Song LI,Zhong-xiao HAO. Trajectory k nearest neighbor query method based on sparse multi-head attention [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1756-1766. |
| [9] | Ming-hui SUN,Hao XUE,Yu-bo JIN,Wei-dong QU,Gui-he QIN. Video saliency prediction with collective spatio-temporal attention [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1767-1776. |
| [10] | Yan-feng LI,Ming-yang LIU,Jia-ming HU,Hua-dong SUN,Jie-yu MENG,Ao-ying WANG,Han-yue ZHANG,Hua-min YANG,Kai-xu HAN. Infrared and visible image fusion based on gradient transfer and auto-encoder [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1777-1787. |
| [11] | Yu-kai LU,Shuai-ke YUAN,Shu-sheng XIONG,Shao-peng ZHU,Ning ZHANG. High precision detection system for automotive paint defects [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(5): 1205-1213. |
| [12] | Li-ming LIANG,Long-song ZHOU,Jiang YIN,Xiao-qi SHENG. Fusion multi-scale Transformer skin lesion segmentation algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(4): 1086-1098. |
| [13] | Yun-zuo ZHANG,Wei GUO,Wen-bo LI. Omnidirectional accurate detection algorithm for dense small objects in remote sensing images [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(4): 1105-1113. |
| [14] | Guo-jun YANG,Ya-hui QI,Xiu-ming SHI. Review of bridge crack detection based on digital image technology [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(2): 313-332. |
| [15] | Xiong-fei LI,Zi-xuan SONG,Rui ZHU,Xiao-li ZHANG. Remote sensing change detection model based on multi⁃scale fusion [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(2): 516-523. |
|
||