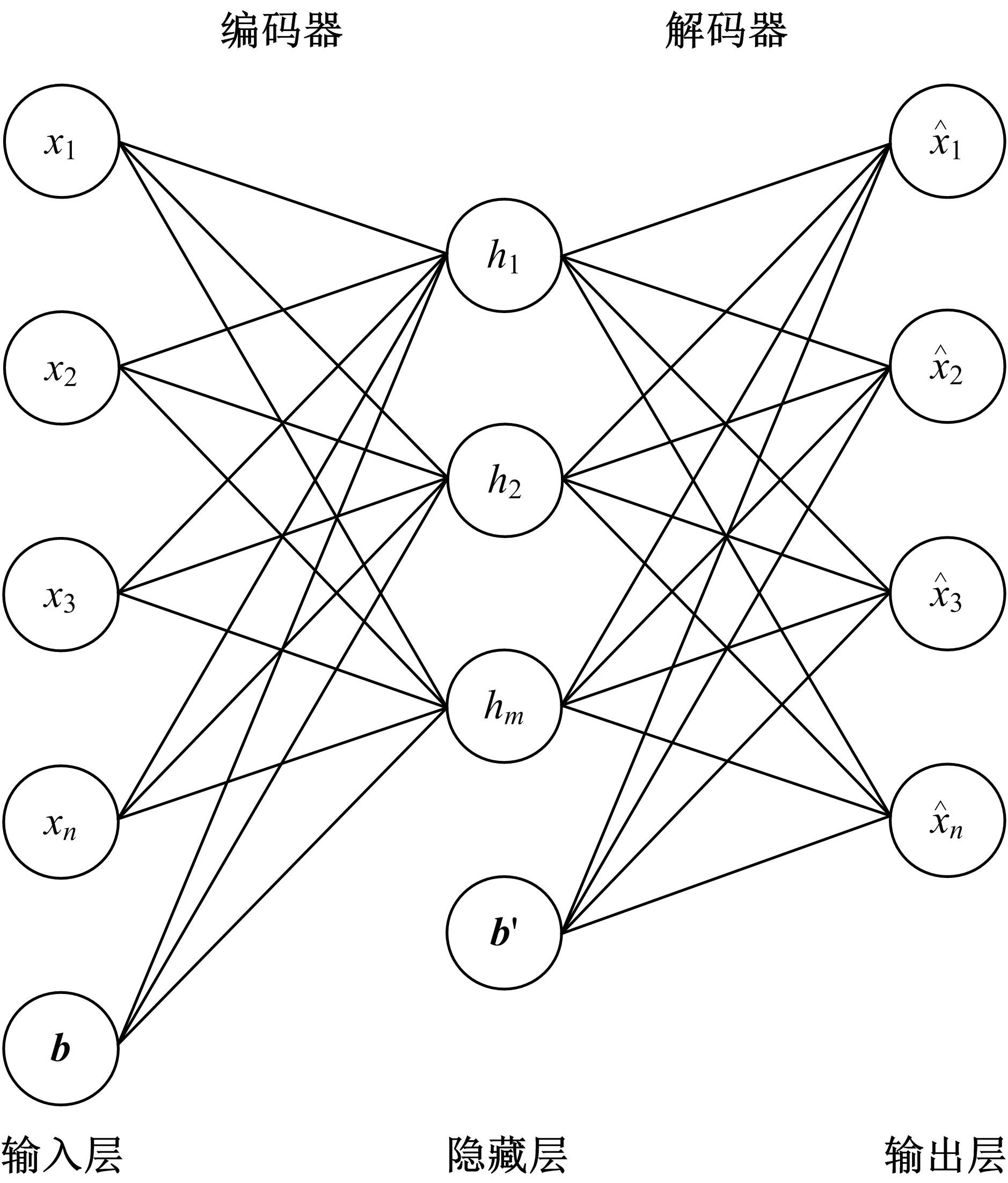
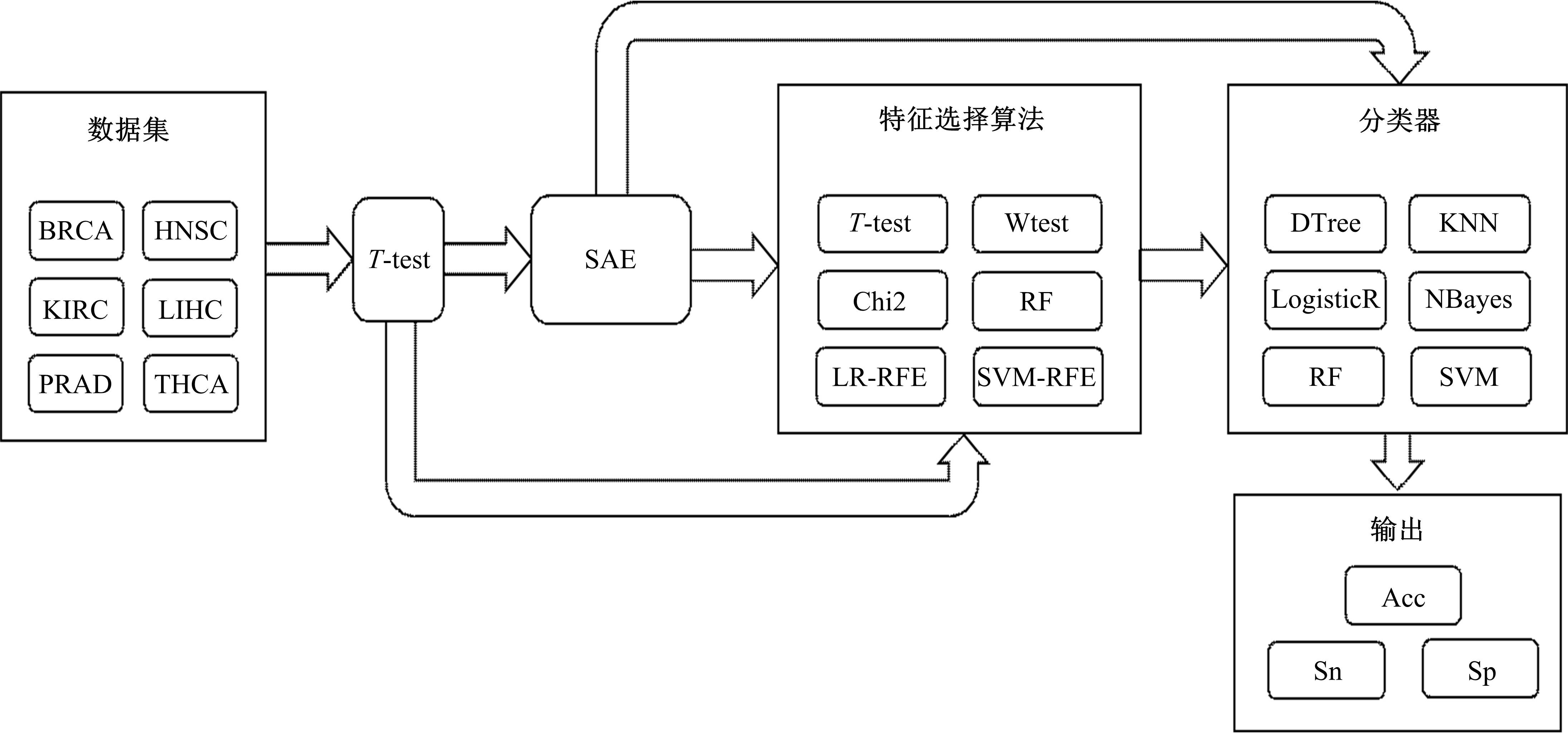
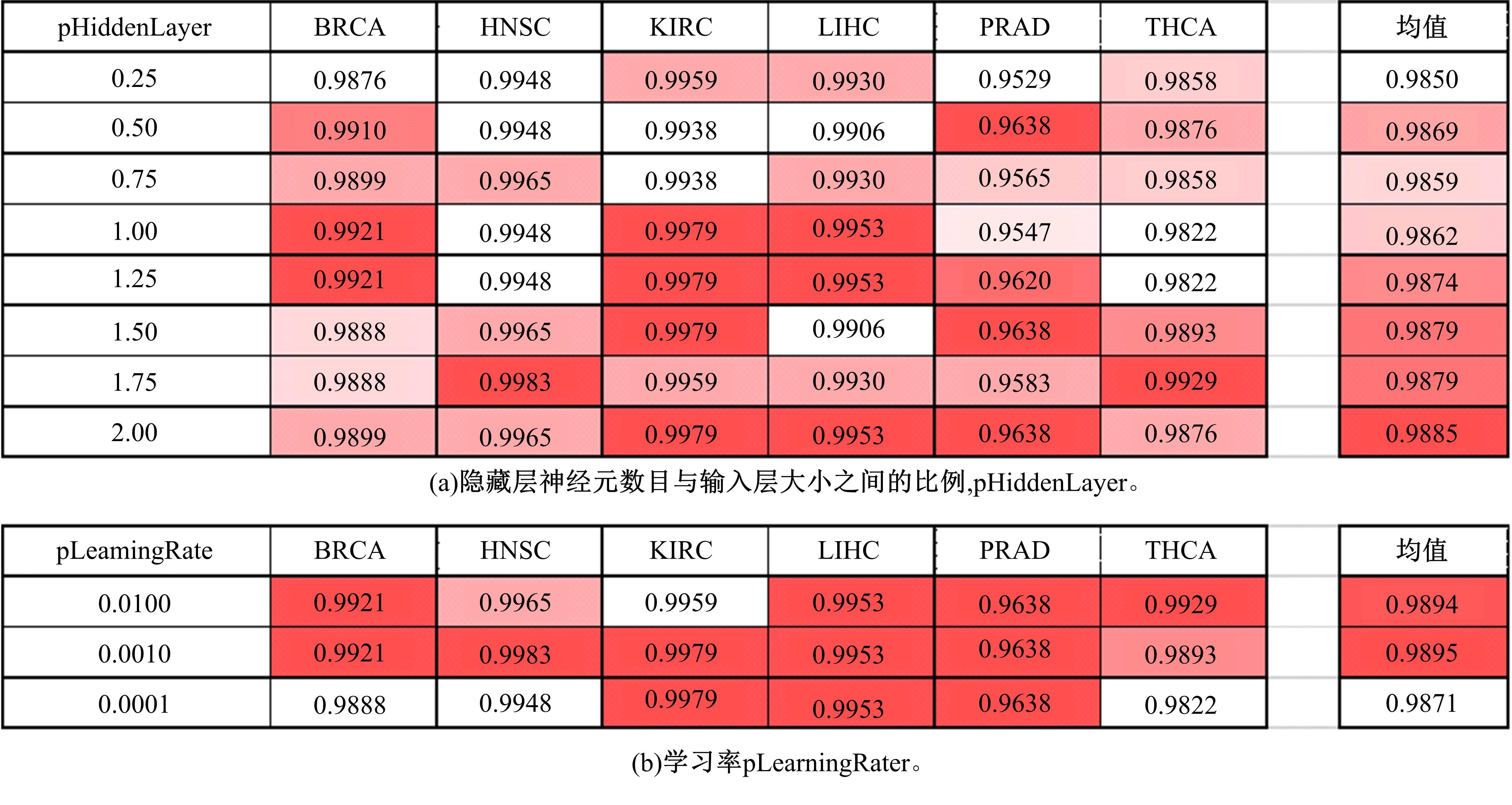
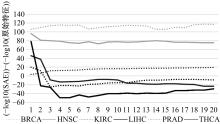
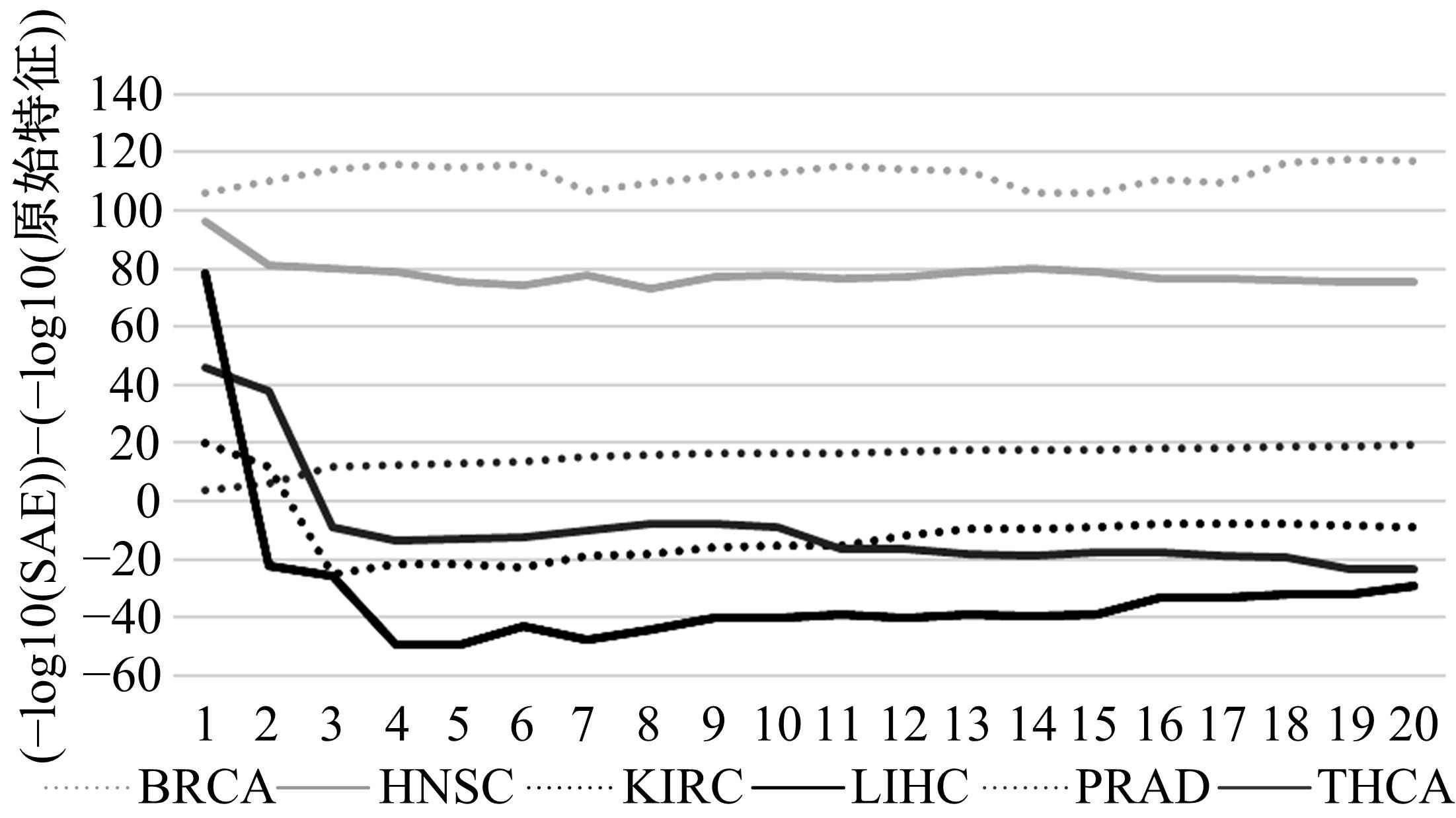
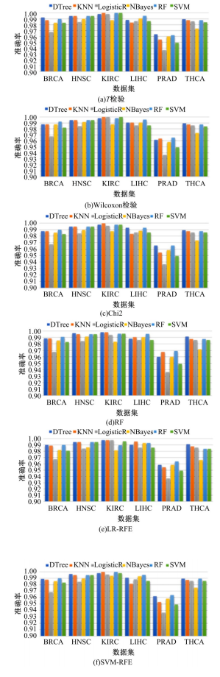
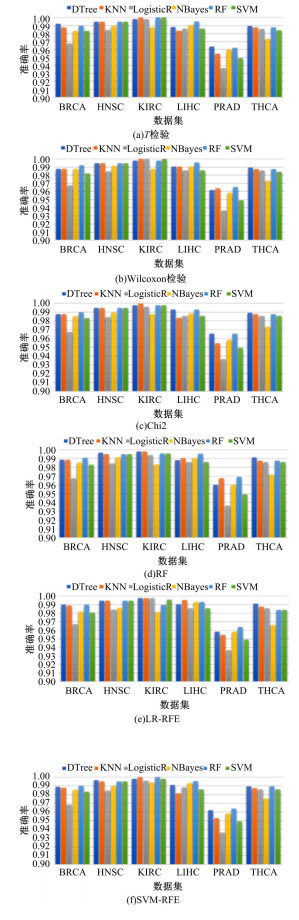
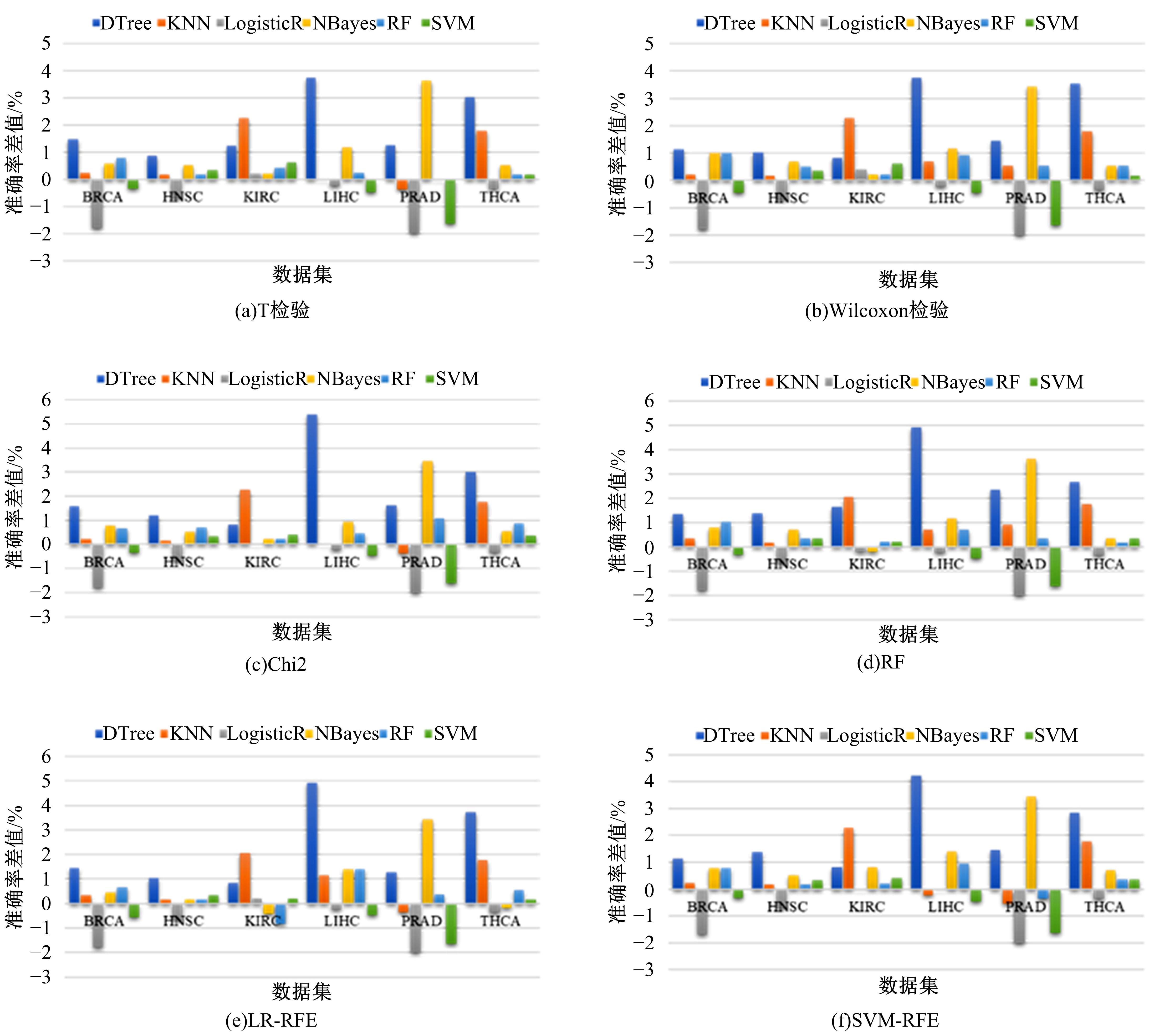
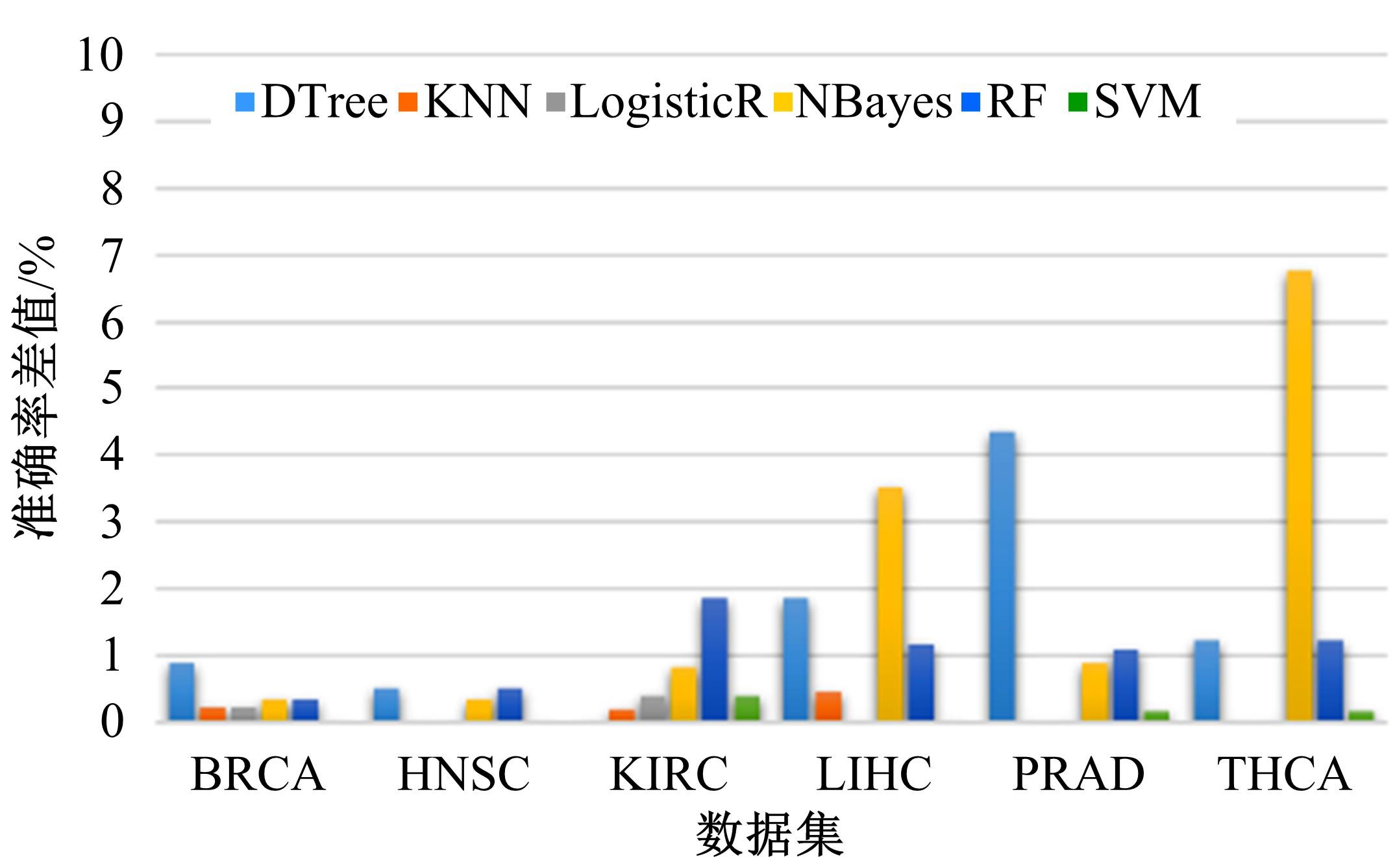
Journal of Jilin University(Engineering and Technology Edition) ›› 2022, Vol. 52 ›› Issue (7): 1645-1656.doi: 10.13229/j.cnki.jdxbgxb20210127
Unsupervised feature engineering algorithm BioSAE based on sparse autoencoder
Feng-feng ZHOU1,2( ),Yi-chi ZHANG1,2
),Yi-chi ZHANG1,2
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education,Jilin University,Changchun 130012,China
CLC Number:
- TP399
| 1 | Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2018, 68(6): 394-424. |
| 2 | Hanahan D, Weinberg R A. Hallmarks of cancer: the next generation[J]. Cell, 2011, 144(5): 646-674. |
| 3 | Martínez-Chantar M L, Avila M A, Lu S C. Hepatocellular carcinoma: updates in pathogenesis, detection and treatment[J]. Cancers, 2020, 12(10): 1-4. |
| 4 | Kandaswamy R, Hannon E, Arseneault L, et al. DNA methylation signatures of adolescent victimization: analysis of a longitudinal monozygotic twin sample[J]. Epigenetics, 2021: 16(11):1169-1186. |
| 5 | Zafon C, Gil J, Pérez-González B, et al. DNA methylation in thyroid cancer[J]. Endocrine-Related Cancer, 2019, 26(7): 415-439. |
| 6 | Bilokapic S, Halic M. Nucleosome and ubiquitin position Set2 to methylate H3K36[J]. Nature Communications, 2019, 10: 1-9. |
| 7 | Champigny M J, Unda F, Skyba O, et al. Learning from methylomes: epigenomic correlates of Populus balsamifera traits based on deep learning models of natural DNA methylation[J]. Plant Biotechnology Journal, 2020, 18(6): 1361-1375. |
| 8 | Levy J J, Titus A J, Petersen C L, et al. MethylNet: an automated and modular deep learning approach for DNA methylation analysis[J]. Bmc Bioinformatics, 2020, 21(1): 1-15. |
| 9 | Zhang M, Pan C, Liu H, et al. An attention-based deep learning method for schizophrenia patients classification using DNA methylation data[C]∥The 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Piscataway, USA, 2020: 172-175. |
| 10 | Shimobaba T, Endo Y, Hirayama R, et al. Autoencoder-based holographic image restoration[J]. Applied Optics, 2017, 56(13): 27-30. |
| 11 | Li F, Zurada J M, Wu W. Sparse representation learning of data by autoencoders with L1/2 regularization[J]. Neural Network, 2018, 28(2): 133-147. |
| 12 | Zhu Y, Qiu P, Ji Y. TCGA-Assembler: open-source software for retrieving and processing TCGA data[J]. Nature Methods, 2014, 11(6): 599-600. |
| 13 | Wei L, Jin Z, Yang S, et al. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data[J]. Bioinformatics, 2018, 34(9): 1615-1617. |
| 14 | Cuturi M. UCI Machine Learning Repository[DB/OL]. [2011-07-24]. . |
| 15 | Diaz-Chito K, Hernández-Sabaté A, López A M. A reduced feature set for driver head pose estimation[J]. Applied Soft Computing, 2016, 45: 98-107. |
| 16 | Abpeikar S, Kasmarik K, Barlow M, et al. UCI machine learning repository[DB/OL]. [2020-06-12]. . |
| 17 | Simon R. Sensitivity, specificity, PPV, and NPV for predictive biomarkers[J]. Journal of the National Cancer Institute, 2015, 107(8): 1-3. |
| 18 | Ghimatgar H, Kazemi K, Helfroush M S, et al. Neonatal EEG sleep stage classification based on deep learning and HMM[J]. Journal of Neural Engineering, 2020, 17(3): 1-17. |
| 19 | Ye Y, Zhang R, Zheng W, et al. RIFS: a randomly restarted incremental feature selection algorithm[J]. Scientific Reports, 2017, 7: 1-11. |
| 20 | 李志军,杨楚皙,刘丹,等. 基于深度卷积神经网络的信息流增强图像压缩方法[J]. 吉林大学学报:工学版, 2020, 50(5): 1788-1795. |
| Li Zhi-jun, Yang Chu-xi, Liu Dan, et al. Deep convolutional networks based image compression with enhancement of information flow[J]. Journal of Jilin University(Engineering and Technology Edition), 2020, 50(5): 1788-1795. | |
| 21 | Xu C, Liu Q, Ye M. Age invariant face recognition and retrieval by coupled auto-encoder networks[J]. Neurocomputing, 2017, 222: 62-71. |
| 22 | Yi B, Shen X, Zhang Z, et al. Expanded autoencoder recommendation framework and its application in movie recommendation[C]∥The 10th International Conference on Software, Knowledge, Information Management & Applications (SKIMA), Piscataway, USA, 2016: 298-303. |
| 23 | Sun W, Shao S, Zhao R, et al. A sparse auto-encoder-based deep neural network approach for induction motor faults classification[J]. Measurement, 2016, 89: 171-178. |
| 24 | 张根保,李浩,冉琰,等. 一种用于轴承故障诊断的迁移学习模型[J]. 吉林大学学报:工学版, 2020, 50(5): 1617-1626. |
| Zhang Gen-bao, Li Hao, Ran Yan, et al. A transfer learning model for bearing fault diagnosis[J]. Journal of Jilin University(Engineering and Technology Edition), 2020, 50(5): 1617-1626. | |
| 25 | Chen Z, Li W. Multisensor feature fusion for bearing fault diagnosis using sparse autoencoder and deep belief network[J]. IEEE Transactions on Instrumentation and Measurement, 2017, 66(7): 1693-1702. |
| 26 | Stirzaker C, Taberlay P C, Statham A L, et al. Mining cancer methylomes: prospects and challenges[J]. Trends in Genetics, 2014, 30(2): 75-84. |
| 27 | Saeed S M U, Anwar S M, Khalid H, et al. EEG based classification of long-term stress using psychological labeling[J]. Sensors, 2020, 20(7): 1-15. |
| 28 | Liao C, Li S, Luo Z. Gene selection for cancer classification using wilcoxon rank sum test and support vector machine[C]∥International Conference on Computational Intelligence and Security, Piscataway, USA, 2007: 368-373. |
| 29 | Feng G, An B, Yang F, et al. Relevance popularity: a term event model based feature selection scheme for text classification[J]. Plos One, 2017, 12(4): 1-15. |
| 30 | Cai J, Xu Y, Zhang W, et al. A comprehensive comparison of residue-level methylation levels with the regression-based gene-level methylation estimations by ReGear[J]. Briefings in Bioinformatics, 2021,22(4): 1-18. |
| 31 | Guyon I, Weston J, Barnhill S, et al. Gene selection for cancer classification using support vector machines[J]. Machine Learning, 2002, 46(1-3): 389-422. |
| 32 | Parikh S A, Gomez R, Thirugnanasambandam M, et al. Decision tree based classification of abdominal aortic aneurysms using geometry quantification measures[J]. Annals of Biomedical Engineering, 2018, 46(12): 2135-2147. |
| 33 | Hu L-Y, Huang M-W, Ke S-W, et al. The distance function effect on k-nearest neighbor classification for medical datasets[J]. Springerplus, 2016, 5(1): 1-9. |
| 34 | Zhu Y, Fang J. Logistic regression-based trichotomous classification tree and its application in medical diagnosis[J]. Medical Decision Making, 2016, 36(8): 973-989. |
| 35 | Ontivero-Ortega M, Lage-Castellanos A, Valente G, et al. Fast gaussian naïve bayes for searchlight classification analysis[J]. Neuroimage, 2017, 163: 471-479. |
| 36 | Sarica A, Cerasa A, Quattrone A. Random forest algorithm for the classification of neuroimaging data in alzheimer's disease: a systematic review[J]. Frontiers in Aging Neuroscience, 2017, 9: 1-12. |
| 37 | Huang S, Cai N, Pacheco P P, et al. Applications of support vector machine (SVM) learning in cancer genomics[J]. Cancer Genomics & Proteomics, 2018, 15(1): 41-51. |
| 38 | Hatfield G W, Hung S P, Baldi P. Differential analysis of DNA microarray gene expression data[J]. Molecular Microbiology, 2003, 47(4): 871-877. |
| 39 | King B H, Robinson C D. Differential analysis of the angle of incidence response of utility-grade PV modules[C]∥The IEEE 46th Photovoltaic Specialists Conference (Pvsc), Piscataway, USA, 2019: 77-81. |
| 40 | Liu T, Wang F, Zhu J, et al. Differential analysis on deep web data sources[C]∥IEEE International Conference on Data Mining Workshops,Piscataway, USA, 2010: 33-40. |
| 41 | Rosenberg T, Kisliouk T, Cramer T, et al. Embryonic heat conditioning induces TET-dependent cross-tolerance to hypothalamic inflammation later in life[J]. Frontiers in Genetics, 2020, 11: 1-20. |
| 42 | Wan P, Long E, Li Z, et al. TET-dependent GDF7 hypomethylation impairs aqueous humor outflow and serves as a potential therapeutic target in glaucoma[J]. Molecular Therapy, 2021, 29: 1-19. |
| [1] | Yao-long KANG,Li-lu FENG,Jing-an ZHANG,Fu CHEN. Outlier mining algorithm for high dimensional categorical data streams based on spectral clustering [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(6): 1422-1427. |
| [2] | Wen-jun WANG,Yin-feng YU. Automatic completion algorithm for missing links in nowledge graph considering data sparsity [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(6): 1428-1433. |
| [3] | Xue-yun CHEN,Xue-yu BEI,Qu YAO,Xin JIN. Pedestrian segmentation and detection in multi-scene based on G-UNet [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 925-933. |
| [4] | Shi-min FANG. Multiple source data selective integration algorithm based on frequent pattern tree [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 885-890. |
| [5] | Da-xiang LI,Meng-si CHEN,Ying LIU. Spontaneous micro-expression recognition based on STA-LSTM [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 897-909. |
| [6] | Ming LIU,Yu-hang YANG,Song-lin ZOU,Zhi-cheng XIAO,Yong-gang ZHANG. Application of enhanced edge detection image algorithm in multi-book recognition [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 891-896. |
| [7] | Xiao-hui WEI,Yan-wei MIAO,Xing-wang WANG. Rhombus sketch: adaptive and more accurate sketch for streaming data [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 874-884. |
| [8] | Xue WANG,Zhan-shan LI,Ying-da LYU. Medical image segmentation based on multi⁃scale context⁃aware and semantic adaptor [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(3): 640-647. |
| [9] | Ji-hong OUYANG,Ze-qi GUO,Si-guang LIU. Dual⁃branch hybrid attention decision net for diabetic retinopathy classification [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(3): 648-656. |
| [10] | Lin MAO,Feng-zhi REN,Da-wei YANG,Ru-bo ZHANG. Two⁃way feature pyramid network for panoptic segmentation [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(3): 657-665. |
| [11] | Xue-zhi WANG,Qing-liang LI,Wen-hui LI. Spatio⁃temporal model of soil moisture prediction integrated with transfer learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(3): 675-683. |
| [12] | Su-ming KANG,Ye-e ZHANG. Hadoop⁃based local timing link prediction algorithm across social networks [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(3): 626-632. |
| [13] | You QU,Wen-hui LI. Single-stage rotated object detection network based on anchor transformation [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(1): 162-173. |
| [14] | Hong-wei ZHAO,Dong-sheng HUO,Jie WANG,Xiao-ning LI. Image classification of insect pests based on saliency detection [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(6): 2174-2181. |
| [15] | Zhou-zhou LIU,Qian-yun ZHANG,Xin-hua MA,Han PENG. Compressed sensing signal reconstruction based on optimized discrete differential evolution algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(6): 2246-2252. |
|
||