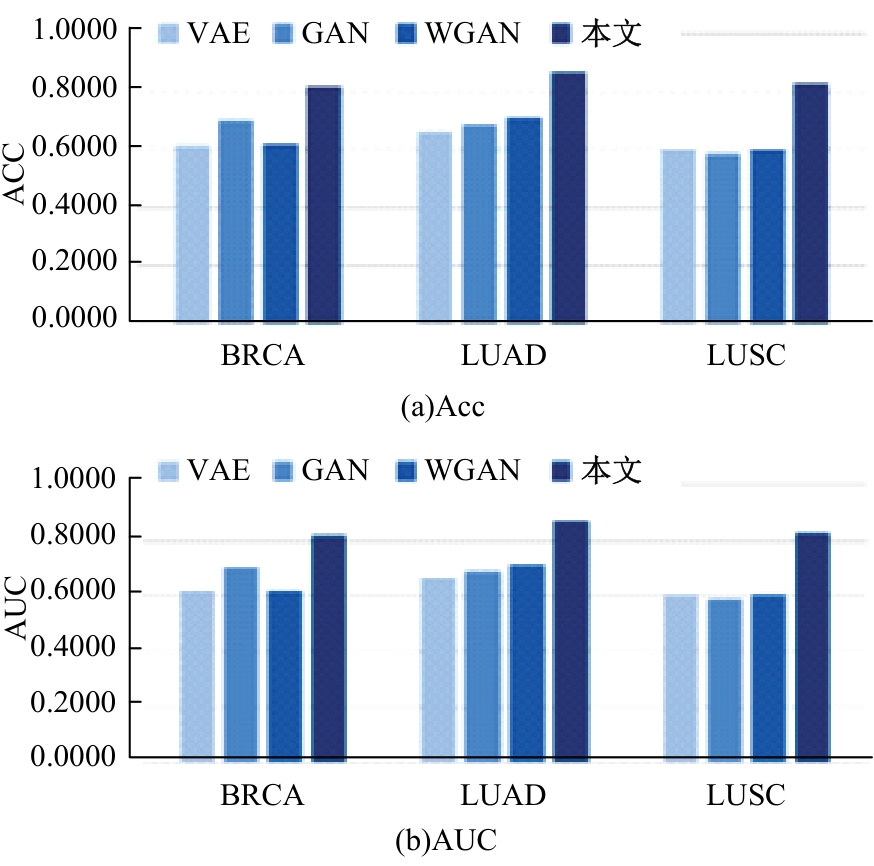
Journal of Jilin University(Engineering and Technology Edition) ›› 2025, Vol. 55 ›› Issue (6): 2089-2096.doi: 10.13229/j.cnki.jdxbgxb.20231196
Previous Articles Next Articles
Feature representation algorithm for imbalanced classification of multi⁃omics cancer data
Feng-feng ZHOU1,2( ),Zhe GUO1,Yu-si FAN2
),Zhe GUO1,Yu-si FAN2
- 1.School of Artificial Intelligence,Jilin University,Changchun 130012,China
2.College of Computer Science and Technology,Jilin University,Changchun 130012,China
CLC Number:
- TP399
| [1] | 安云鹤, 李宝明, 李越, 等. 癌症基因组测序方案制定的研究进展[J]. 中国生物工程杂志, 2014, 34(11): 9-17. |
| An Yun-he, Li Bao-ming, Li Yue, et al. Progress in cancer genome-sequencing study design[J]. China Biotechnology, 2014, 34(11): 9-17. | |
| [2] | 周丰丰, 张亦弛. 基于稀疏自编码器的无监督特征工程算法BioSAE[J].吉林大学学报: 工学版, 2022, 52(7): 1645-1656. |
| Zhou Feng-feng, Zhang Yi-chi. Unsupervised feature engineering algorithm BioSAE based on sparse autoencoder[J]. Journal of Jilin University (Engineering and Technology Edition), 2022, 52(7): 1645-1656. | |
| [3] | Chen X Y, Yu Y Z W, Zheng H Y, et al. Single-cell transcriptome analysis reveals dynamic changes of the preclinical A549 cancer models, and the mechanism of dacomitinib[J]. European Journal of Pharmacology, 2023, 960: No.176046. |
| [4] | 白天, 周春光, 王喆, 等. 代谢组学中机器学习研究进展[J]. 吉林大学学报: 信息科学版, 2008, 26(2): 163-168. |
| Bai Tian, Zhou Chun-guang, Wang Zhe, et al. Advances of machine learning in metabonomics[J]. Journal of Jilin University (Information Science Edition), 2008, 26(2): 163-168. | |
| [5] | 高美虹, 尚学群. 利用人工智能预测癌症的易感性、复发性和生存期[J]. 生物化学与生物物理进展, 2022, 49(9): 1687-1702. |
| Gao Mei-hong, Shang Xue-qun. Artificial intelligence-based prediction for cancer susceptibility, recurrence and survival[J]. Progress in Biochemistry and Biophysics, 2022, 49(9): 1687-1702. | |
| [6] | 刘富, 梁艺馨, 侯涛, 等. 模糊c-harmonic均值算法在不平衡数据上改进[J]. 吉林大学学报: 工学版, 2021, 51(4): 1447-1453. |
| Liu Fu, Liang Yi-xin, Hou Tao, et al. Improvement of fuzzy c-harmonic mean algorithm on unbalanced data[J]. Journal of Jilin University (Engineering and Technology Edition), 2021, 51(4): 1447-1453. | |
| [7] | 章鸯, 潘飞燕, 章卫国, 等. 高通量测序在无创产前遗传学诊断中的应用价值[J]. 中国卫生检验杂志, 2022, 32(10): 1249-1253. |
| Zhang Yang, Pan Fei-yan, Zhang Wei-guo, et al. Application value of high -throughput sequencing noninvasive prenatal testing in prenatal genetic diagnosis[J]. Chin J Health Lab Tec, 2022, 32(10): 1249-1253. | |
| [8] | 方朝剑, 胡新荣. 基于模糊近似度的隐私敏感数据过滤算法[J]. 吉林大学学报: 工学版, 2023, 53(4): 1174-1180. |
| Fang Chao-jian, Hu Xin-rong. Privacy-sensitive data filtering algorithm based on fuzzy approximation[J]. Journal of Jilin University (Engineering and Technology Edition), 2023, 53(4): 1174-1180. | |
| [9] | 张浩, 李海鹏, 彭国琴, 等. 多层次特征融合表征的图像情感识别[J]. 计算机辅助设计与图形学学报, 2023, 35: 1-11. |
| Zhang Hao, Li Hai-peng, Peng Guo-qin, et al. Image emotion recognition via fusion multi-level representations[J]. Journal of Computer-Aided Design & Computer Graphics, 2023, 35: 1-11. | |
| [10] | Alain L U, Melissa S, Péter O, et al. Transcriptome-based identification of novel endotypes in adult atopic dermatitis[J]. Allergy, 2022, 77(5): 1486-1498. |
| [11] | Sun M, Ding T, Tang X Q, et al. An efficient mixed-model for screening differentially expressed genes of breast cancer based on LR-RF[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2019, 16(1): 124-130. |
| [12] | Ball M W, Gorin M A, Drake C G, et al. The landscape of whole-genome alterations and pathologic features in genitourinary malignancies: An analysis of the cancer genome atlas[J]. European Urology Focus, 2017, 3(6): 584-589. |
| [13] | Bourgade R, Rabilloud N, Perennec T, et al. Deep learning for detecting BRCA mutations in high-grade ovarian cancer based on an innovative tumor segmentation method from whole slide images[J]. Modern Pathology, 2023, 36(11): No.100304. |
| [14] | Yildirimtepe C F, Ercan C. RGS10 suppression by DNA methylation is associated with low survival rates in colorectal carcinoma[J]. Pathology - Research and Practice, 2022, 236: No.154007. |
| [15] | Aysegul C, Cenk A A, Yalcin A K. Novel molecular signatures and potential therapeutics in renal cell carcinomas: Insights from a comparative analysis of subtypes[J]. Genomics, 2020, 112(5): 3166-3178. |
| [16] | Turab N A A, Murtaza R S A, Imtaiyaz H M. Pan-cancer analysis of Chromobox (CBX) genes for prognostic significance and cancer classification[J]. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 2023, 1869(1): No.166561. |
| [17] | Goodfellow I J, Jean P A, Mehdi M, et al. Generative adversarial nets[C]∥Proceedings of the 27th International Conference on Neural Information Processing Systems, Montreal, Canada, 2014: 2672-2680. |
| [18] | Kingma D P. Welling M. Auto-encoding variational Bayes[J/OL].[2023-10-22]. |
| [19] | Precup D, Teh Y W. Wasserstein Generative Adversarial Networks[C]∥Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 2017, 70: 214-223. |
| [20] | Chawla N V, Bowyer K W, Hall L O, et al. SMOTE: synthetic minority over-sampling technique[J]. J. Artif. Int. Res.,2002, 16(1): 321-357. |
| [21] | 周丰丰, 孙燕杰, 范雨思. 基于miRNA组学的数据增强算法[J]. 电子科技大学学报, 2023, 52(2): 182-187. |
| Zhou Feng-feng, Sun Yan-jie, Fan Yu-si. Data augmentation algorithm for miRNA omics-based classifications[J]. Journal of University of Electronic Science and Technology of China, 2023, 52(2): 182-187. |
| [1] | Jian WANG,Chen-wei JIA. Trajectory prediction model for intelligent connected vehicle [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(6): 1963-1972. |
| [2] | Xiang-jiu CHE,Yu-peng SUN. Graph node classification algorithm based on similarity random walk aggregation [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(6): 2069-2075. |
| [3] | Xiu-zhi ZHAO,De-hong XIE. Discrimination method for Pu-er tea varieties based on noise-robust feature extraction [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1756-1762. |
| [4] | Qing-liang JIN,Xin-sen ZHOU,Yi CHEN,Cheng-wen WU. Predictive model for identifying innovative university talents based on the swarm intelligence evolution enhanced kernel extreme learning machine [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1763-1771. |
| [5] | Sheng-qi MEI,Xiao-dong LIU,Xing-ju WANG,Xu-feng LI,Teng WU,Xiang-xu CHENG. Prediction of high strength concrete creep based on parametric MIC analysis and machine learning algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1595-1603. |
| [6] | Jun WANG,Chang-fu SI,Kai-peng WANG,Qiang FU. Intrusion detection method based on ensemble learning and feature selection by PSO-GA [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(4): 1396-1405. |
| [7] | Xiang-hai MENG,Guo-rui WANG,Ming-yang ZHANG,Bi-jiang TIAN. Traffic accident prediction model of mountain highways based on selection integration [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(4): 1298-1306. |
| [8] | Yin-fei DAI,Xiu-zhen ZHOU,Yu-bao LIU,Zhi-yuan LIU. In⁃vehicle network intrusion detection system based on CAN bus data [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(3): 857-865. |
| [9] | Xiang-jiu CHE,Yu-ning WU,Quan-le LIU. A weighted isomorphic graph classification algorithm based on causal feature learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(2): 681-686. |
| [10] | Lei ZHANG,Jing JIAO,Bo-xin LI,Yan-jie ZHOU. Large capacity semi structured data extraction algorithm combining machine learning and deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(9): 2631-2637. |
| [11] | Cheng CHEN,Pei-xin SHI,Peng-jiao JIA,Man-man DONG. Correlation analysis of shield driving parameters and structural deformation prediction based on MK-LSTM algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1624-1633. |
| [12] | Li-ming LIANG,Long-song ZHOU,Jiang YIN,Xiao-qi SHENG. Fusion multi-scale Transformer skin lesion segmentation algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(4): 1086-1098. |
| [13] | Bo-song FAN,Chun-fu SHAO. Urban rail transit emergency risk level identification method [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(2): 427-435. |
| [14] | Shi-feng NIU,Shi-jie YU,Yan-jun LIU,Chong MA. Real-time detection method of angry driving behavior based on bracelet data [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(12): 3505-3512. |
| [15] | Dondrub LHAKPA,Duoji ZHAXI,Jie ZHU. Tibetan text normalization method [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(12): 3577-3588. |
|