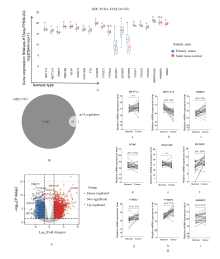
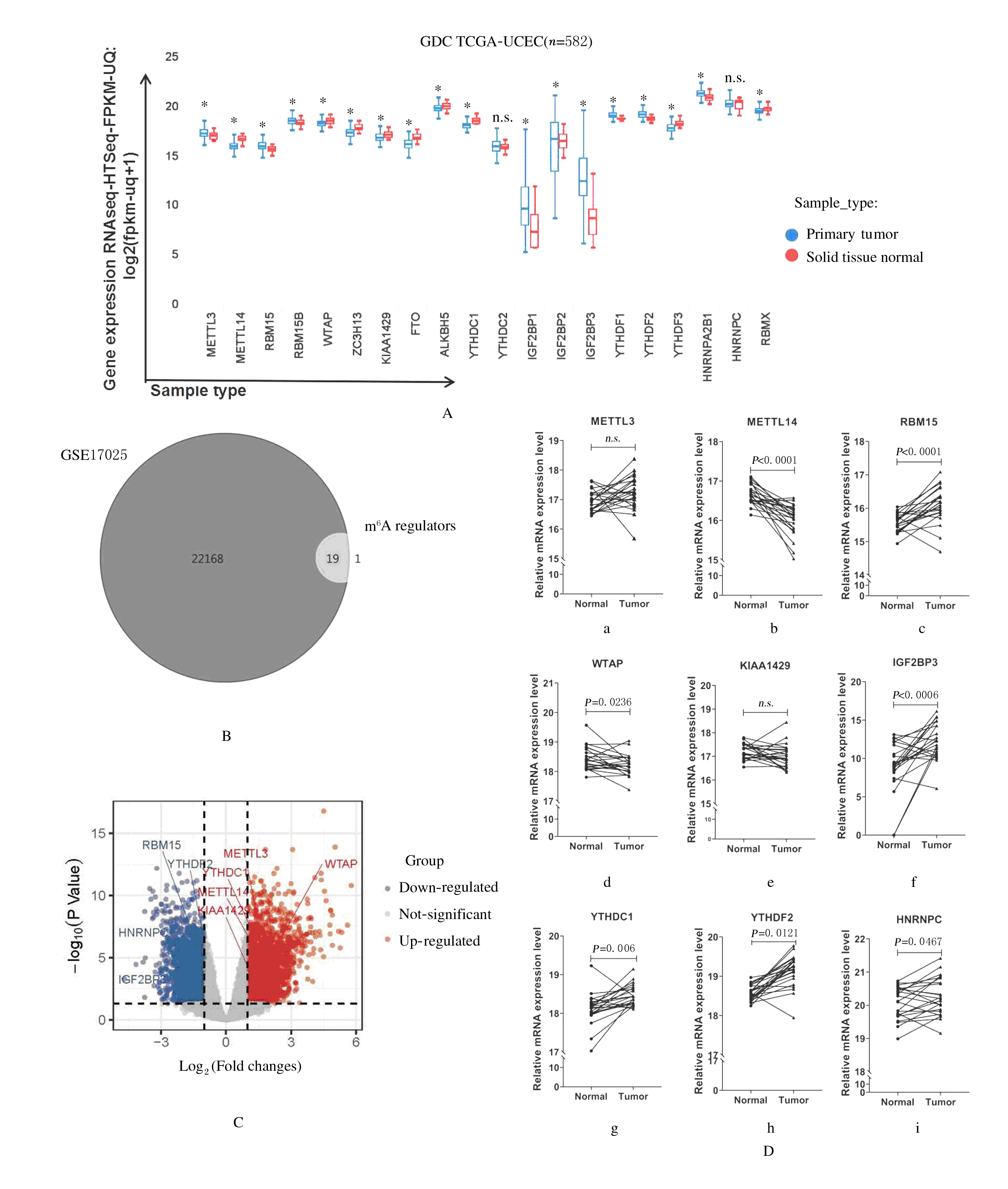
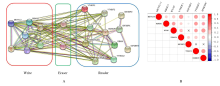
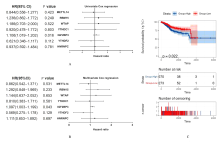
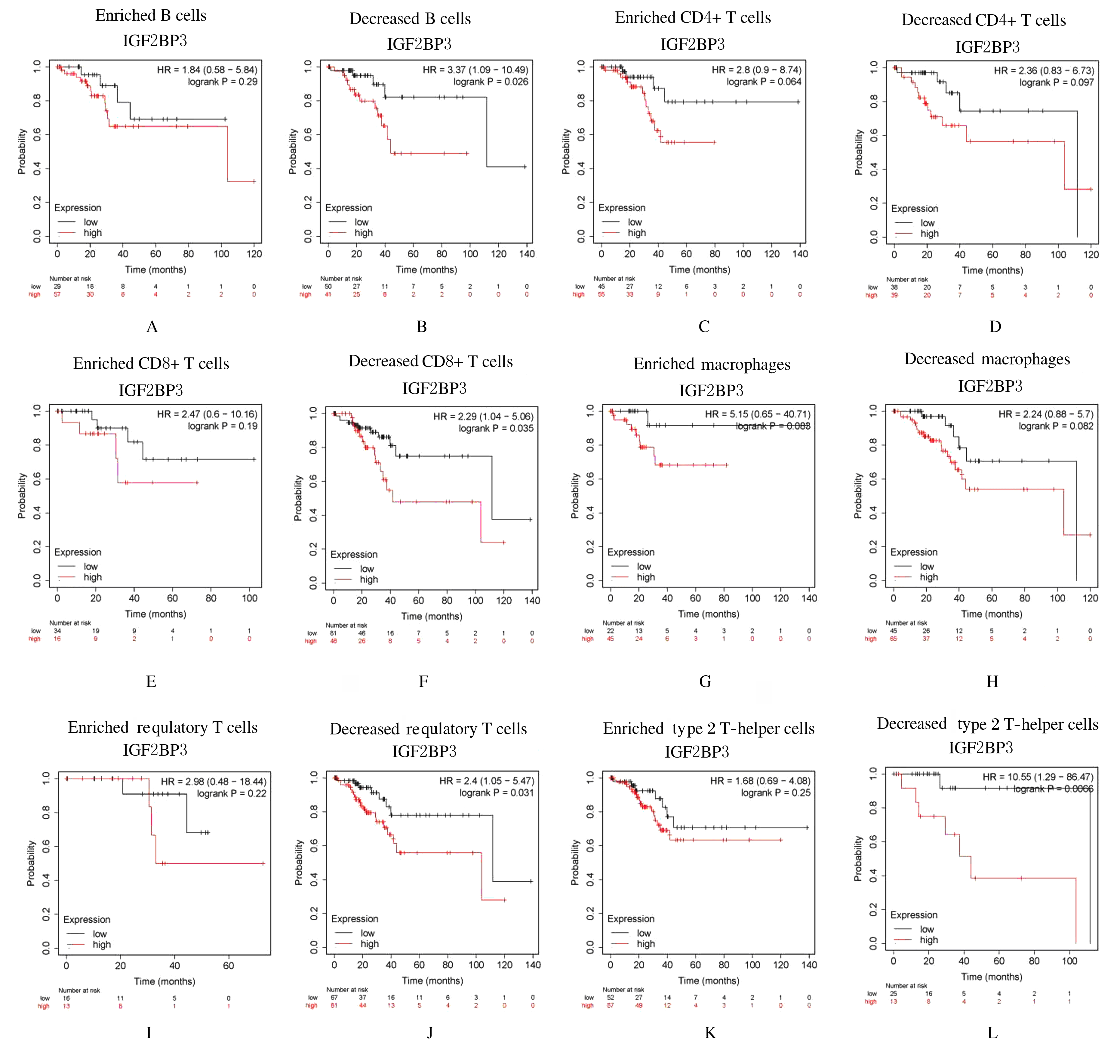
| 1 |
LU K, BROADDUS R. Endometrial cancer [J]. N Engl Med, 2020, 383(21): 2053-2064.
|
| 2 |
DANAHER P, WARREN S, LU R, et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA) [J]. Immunother Cancer, 2018, 6(1): 63.
|
| 3 |
VANDERSTRAETEN A, TUYAERTS S, AMANT F. The immune system in the normal endometrium and implications for endometrial cancer development [J]. Reproductive Immunol, 2015, 109: 7-16.
|
| 4 |
GUO F, DONG Y, TAN Q, et al. Tissue infiltrating immune cells as prognostic biomarkers in endometrial cancer: A Meta-analysis [J]. Dis Markers, 2020, 2020: 1805764.
|
| 5 |
DE FELICE F, MARCHETTI C, TOMBOLINI V, et al. Immune check-point in endometrial cancer [J]. Int J Clin Oncol, 2019, 24(8): 910-916.
|
| 6 |
VERSLUIS M, MARCHAL S, PLAT A, et al. The prognostic benefit of tumour-infiltrating Natural Killer cells in endometrial cancer is dependent on concurrent overexpression of human leucocyte antigen-E in the tumour microenvironment [J]. Eur J Cancer, 2017, 86: 285-295.
|
| 7 |
WORKEL H, KOMDEUR F, WOUTERS M, et al. CD103 defines intraepithelial CD8+ PD1+ tumour-infiltrating lymphocytes of prognostic significance in endometrial adenocarcinoma [J]. Eur J Cancer, 2016, 60: 1-11.
|
| 8 |
LAN Q, LIU P, HAASE J, et al. The critical role of RNA m6A methylation in cancer [J]. Cancer Res, 2019, 79(7): 1285-1292.
|
| 9 |
WANG Y Z, REN F, SONG Z X, et al. Multiomics profile and prognostic gene signature of m6A regulators in uterine corpus endometrial carcinoma [J]. Cancer, 2020, 11(21): 6390-6401.
|
| 10 |
SHULMAN Z, STERN-GINOSSAR N. The RNA modification N6-methyladenosine as a novel regulator of the immune system [J]. Nat Immunol, 2020, 21(5): 501-512.
|
| 11 |
YI L, WU G, GUO L, et al. Comprehensive analysis of the PD-L1 and immune infiltrates of mA RNA methylation regulators in head and neck squamous cell carcinoma [J]. Mol Ther Nucleic Acids, 2020, 21: 299-314.
|
| 12 |
WU H, DONG H, FU Y, et al. Expressions of m6A RNA methylation regulators and their clinical predictive value in cervical squamous cell carcinoma and endometrial adenocarcinoma [J]. Clin Exp Pharmacol Physiol, 2021, 48(2): 270-278.
|
| 13 |
DANAHER P, WARREN S, DENNIS L, et al. Gene expression markers of tumor infiltrating leukocytes [J]. Immunother Cancer, 2017, 5: 18.
|
| 14 |
SOUSA S, MÄÄTTÄ J. The role of tumour-associated macrophages in bone metastasis [J]. Bone Oncol, 2016, 5(3): 135-138.
|
| 15 |
SIEMERS N, HOLLOWAY J, CHANG H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors [J]. PLoS One, 2017, 12(7): e0179726.
|
| 16 |
YANG J, LI H X, HU S D, et al. ACE2 correlated with immune infiltration serves as a prognostic biomarker in endometrial carcinoma and renal papillary cell carcinoma: implication for COVID-19 [J]. Aging, 2020, 12(8): 6518-6535.
|
| 17 |
SCHOBER P, BOER C, SCHWARTE L. Correlation coefficients: appropriate use and interpretation [J]. Anesth Analg, 2018, 126(5): 1763-1768.
|
| 18 |
NETWORK C G A R, KANDOTH C,SCHULTZ N, et al. Integrated genomic characterization of endometrial carcinoma [J]. Nature, 2013, 497(7447): 67-73.
|
| 19 |
PAKISH J B, ZHANG Q, CHEN Z Y, et al. Immune microenvironment in microsatellite-instable endometrial cancers: hereditary or sporadic origin matters [J]. Clin Cancer Res, 2017, 23(15): 4473-4481.
|
| 20 |
LI C, ZOTA V, WODA B, et al. Expression of a novel oncofetal mRNA-binding protein IMP3 in endometrial carcinomas: diagnostic significance and clinicopathologic correlations [J]. Mod Pathol, 2007, 20(12): 1263-1268.
|
| 21 |
ZHENG W X, YI X F, FADARE O, et al. The oncofetal protein IMP3: a novel biomarker for endometrial serous carcinoma [J]. Am Surg Pathol, 2008, 32(2): 304-315.
|
| 22 |
VISSER N, PUTTEN LVAN D E R, EGERSCHOT A V A N, et al. Addition of IMP3 to L1CAM for discrimination between low- and high-grade endometrial carcinomas: a European Network for Individualised Treatment of Endometrial Cancer collaboration study[J]. Hum Pathol, 2019, 89(7): 90-98.
|
| 23 |
MHAWECH-FAUCEGLIA P, HERRMANN F R, RAI H, et al. IMP3 distinguishes uterine serous carcinoma from endometrial endometrioid adenocarcinoma [J]. Am J Clin Pathol, 2010, 133(6): 899-908.
|
| 24 |
LI W, LIU D, CHANG W, et al. Role of IGF2BP3 in trophoblast cell invasion and migration [J]. Cell Death Dis, 2014, 5: e1025.
|
| 25 |
HAN D L, LIU J Y, CHEN C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells [J].Nature, 2019,566(7743): 270-274.
|
| 26 |
ZHANG B, WU Q, LI B, et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer [J]. Mol Cancer, 2020, 19(1): 53.
|
| 27 |
ZHENG Q, HOU J, ZHOU Y, et al. The RNA helicase DDX46 inhibits innate immunity by entrapping mA-demethylated antiviral transcripts in the nucleus [J]. Nat Immunol, 2017, 18(10): 1094-1103.
|
| 28 |
WINKLER R, GILLIS E, LASMAN L, et al. m6A modification controls the innate immune response to infection by targeting type Ⅰ interferons [J]. Nat Immunol, 2019, 20(2): 173-182.
|
| 29 |
RUBIO R M, DEPLEDGE D P, BIANCO C, et al. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon B [J]. Genes Develop, 2018, 32: 1472-1484.
|
| 30 |
LI H B, TONG J Y, ZHU S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways [J]. Nature, 2017, 548(7667): 338-342.
|
 ),杨洋1(
),杨洋1( )
)
 ),Yang YANG1(
),Yang YANG1( )
)