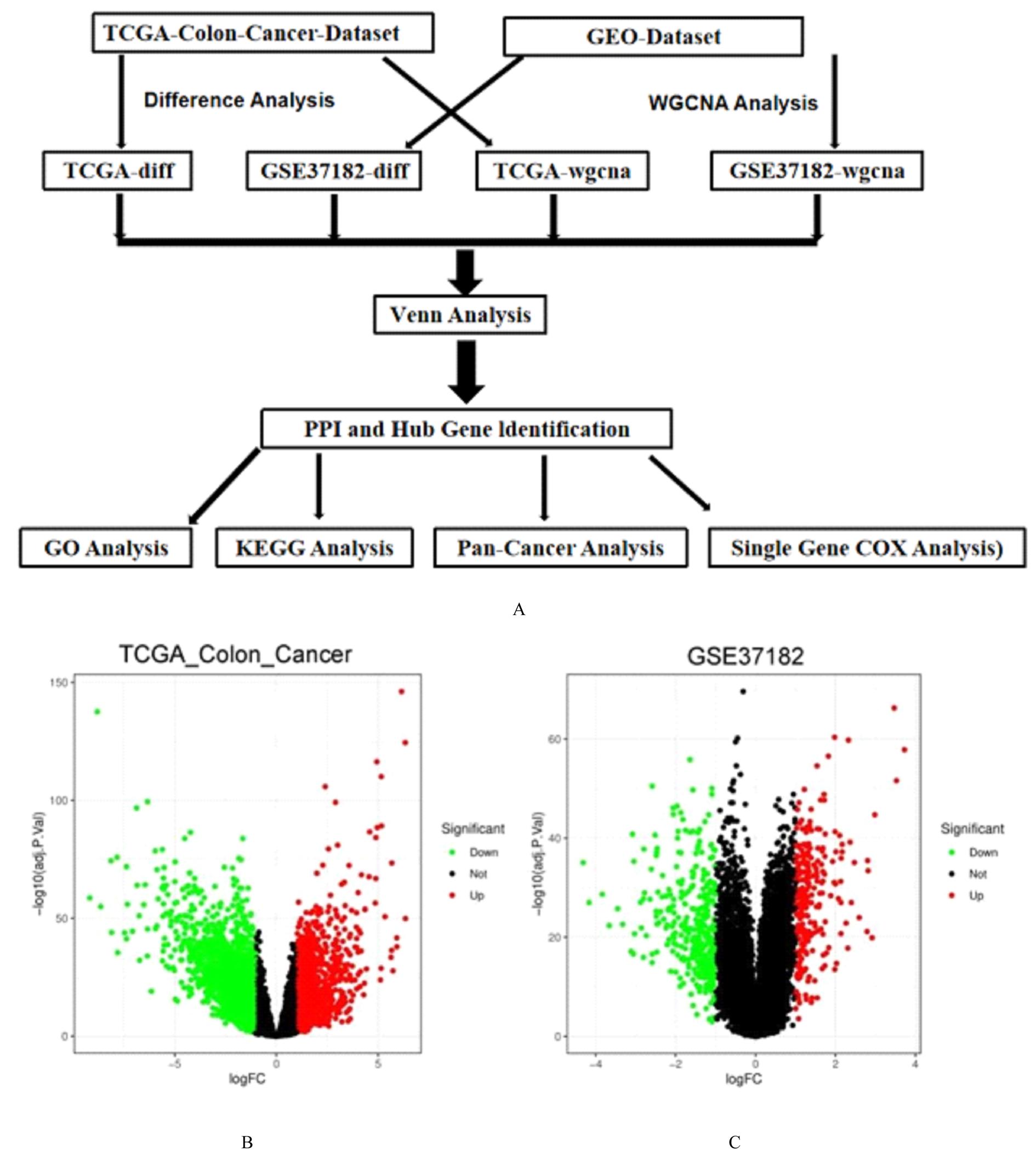
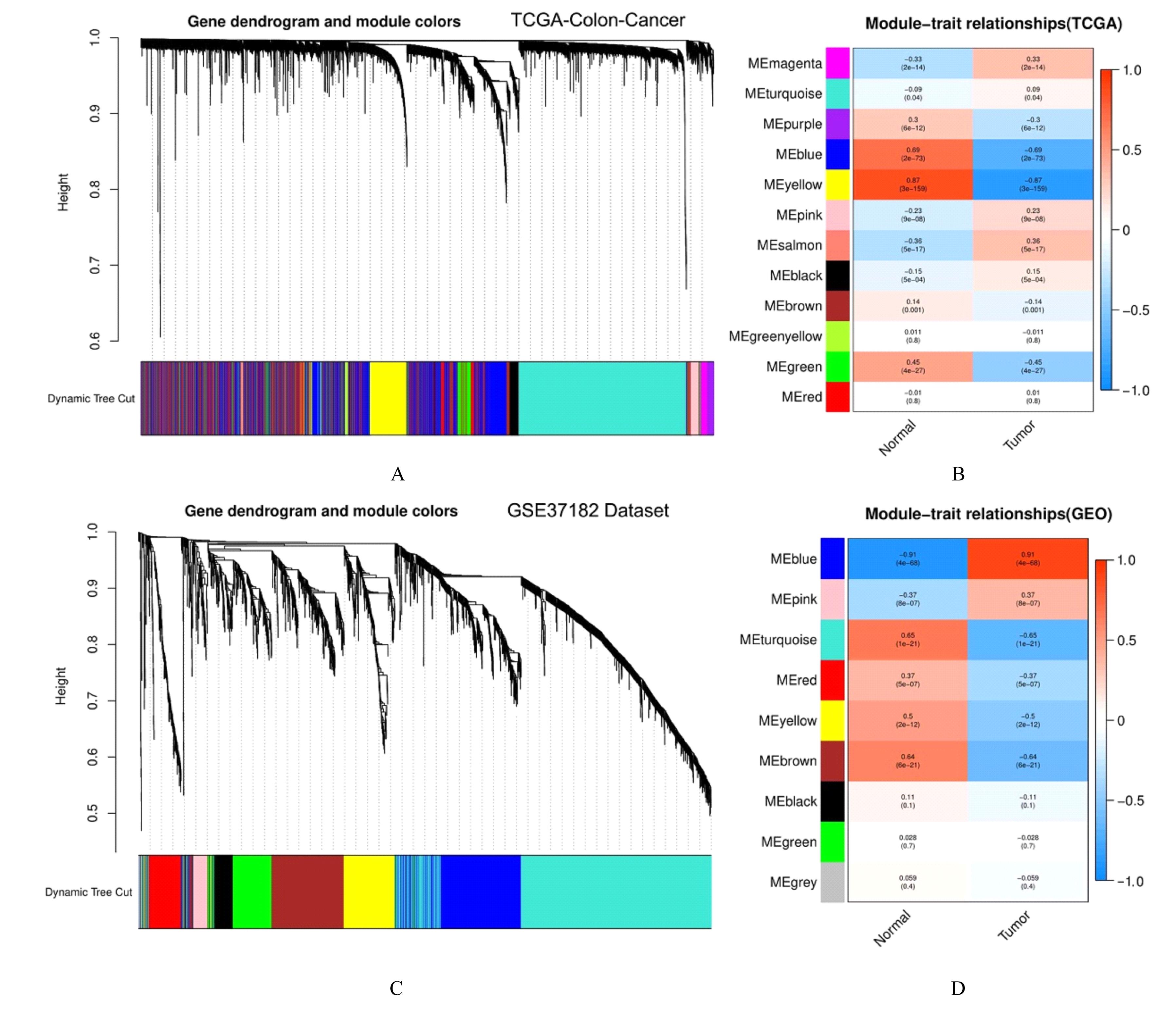
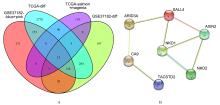
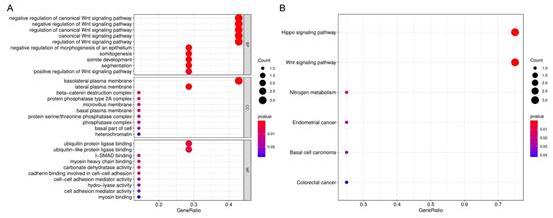
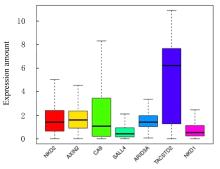
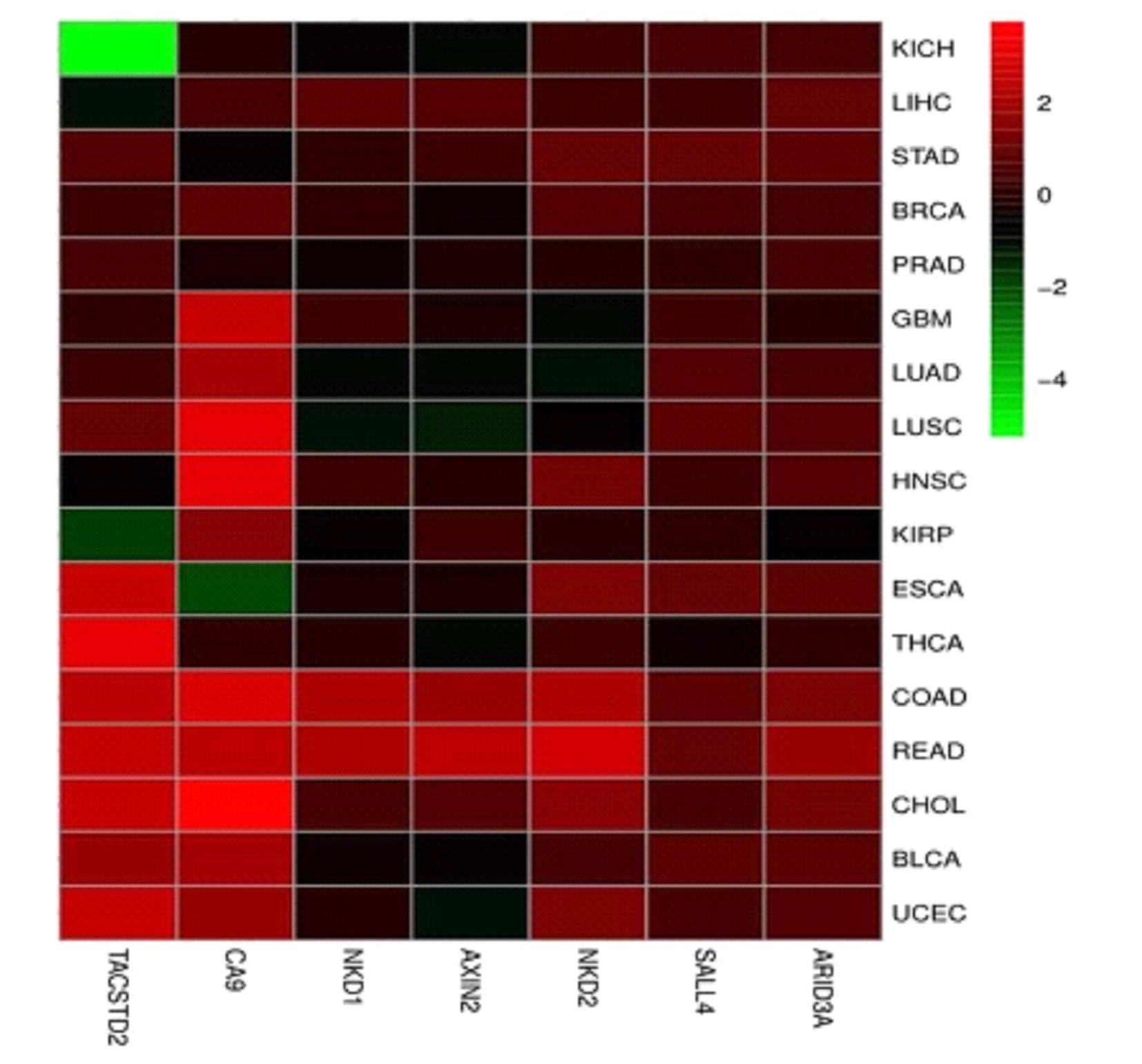
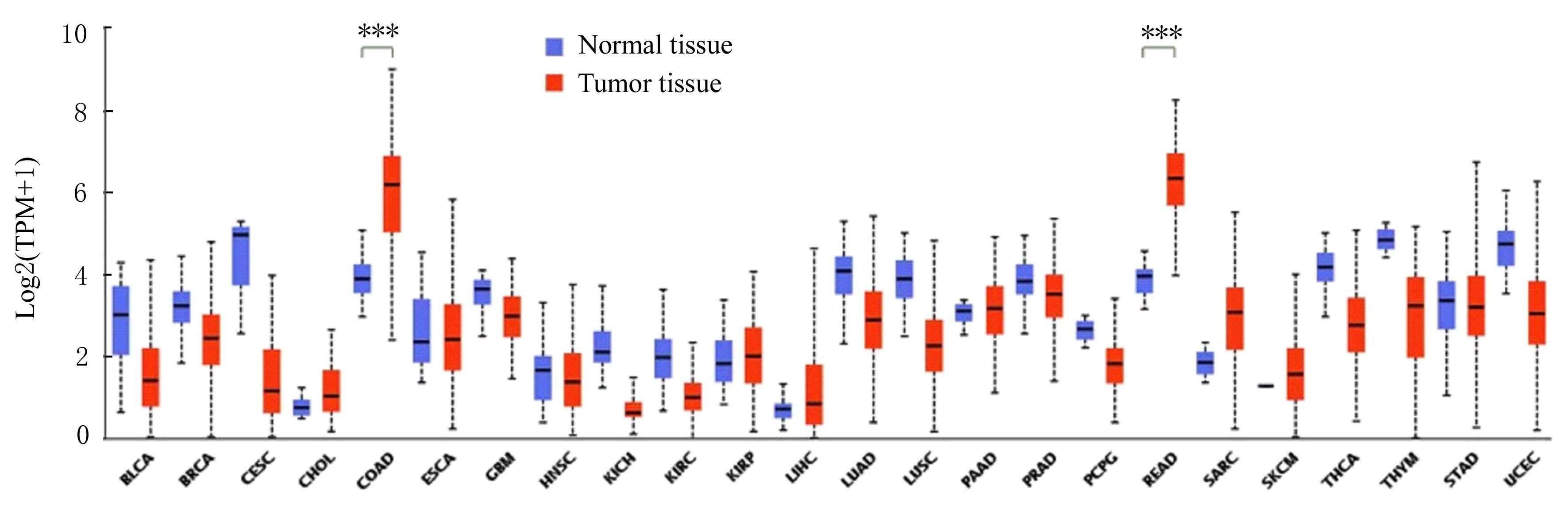
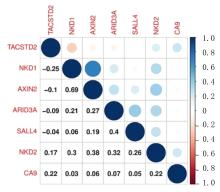
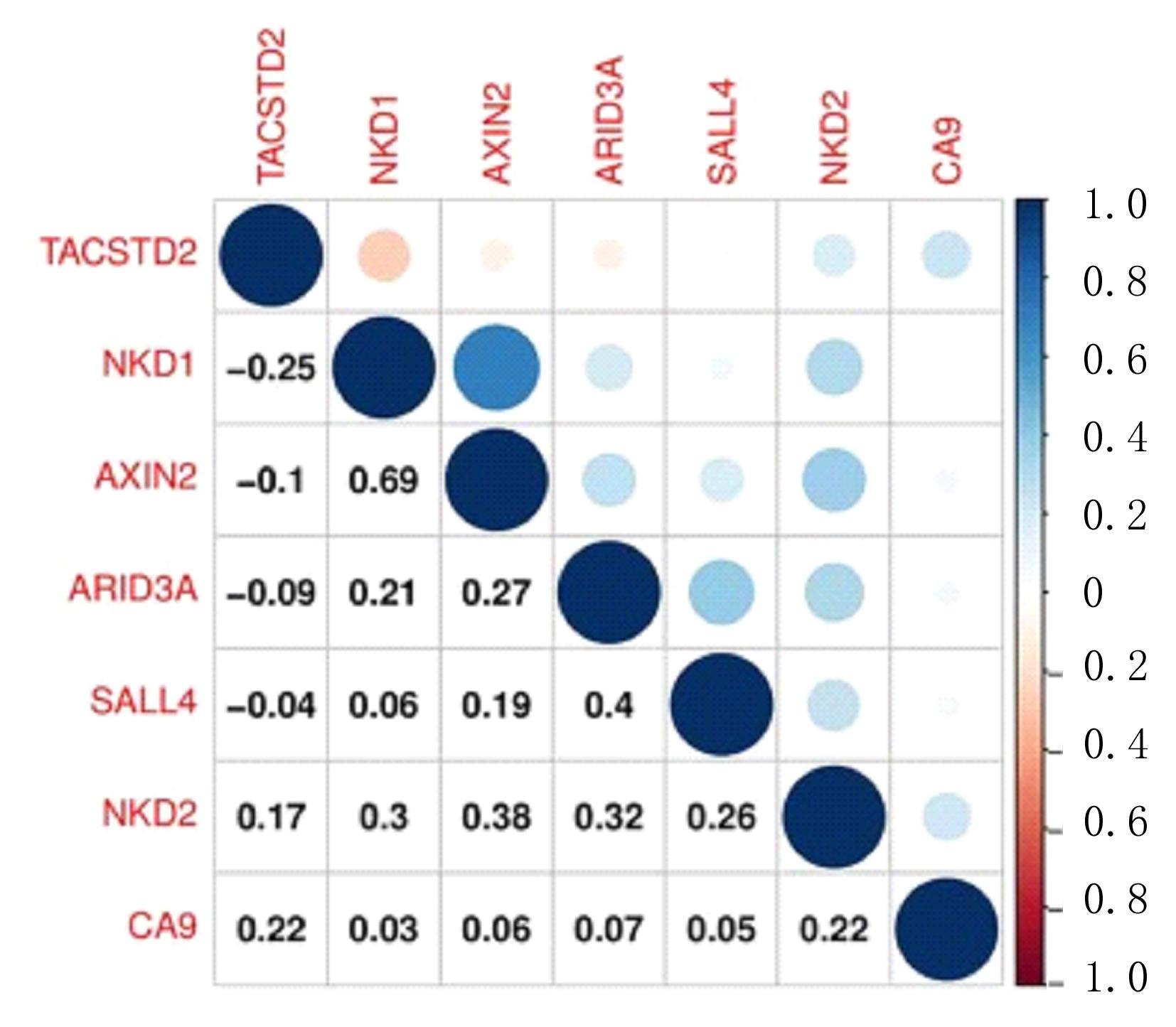
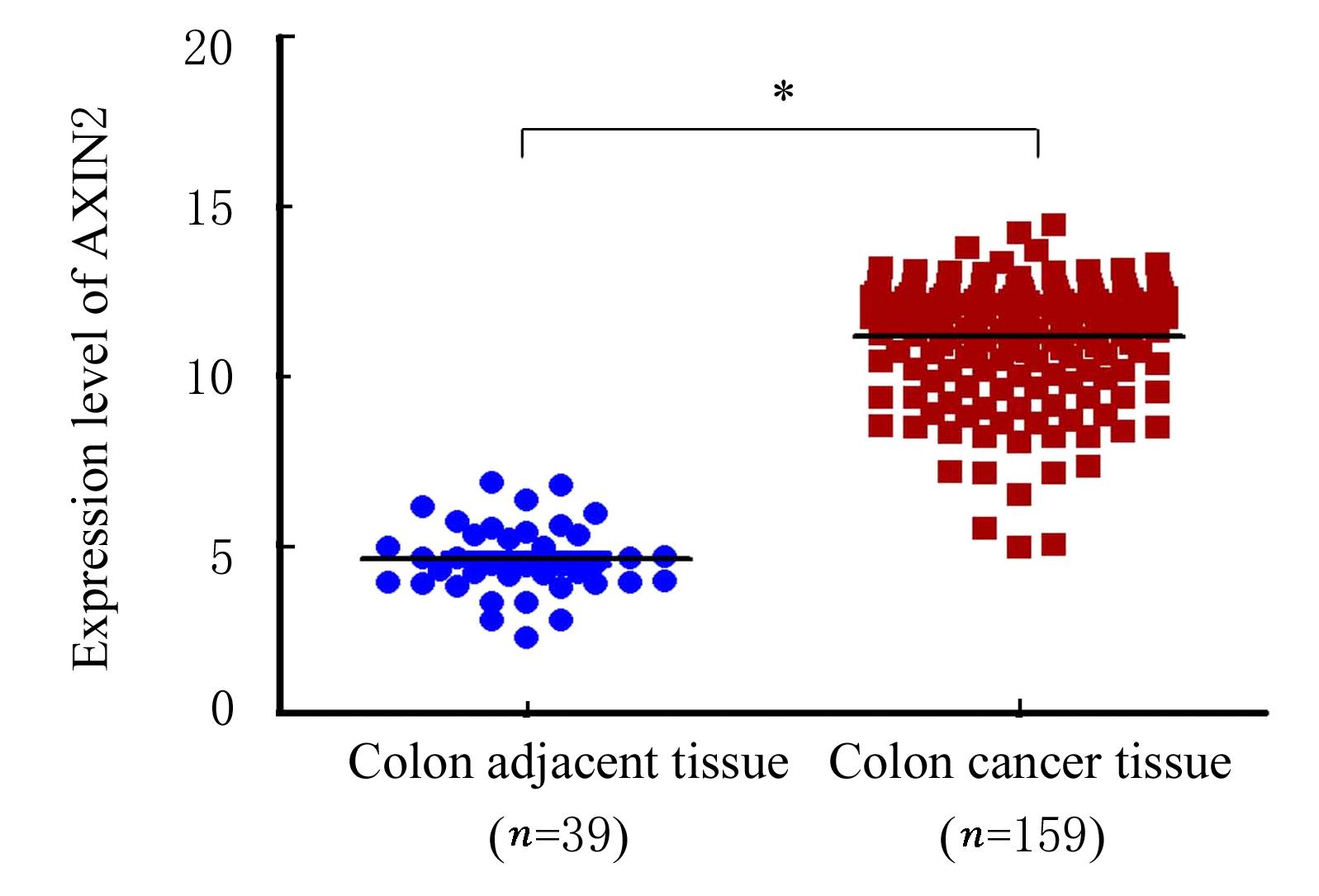
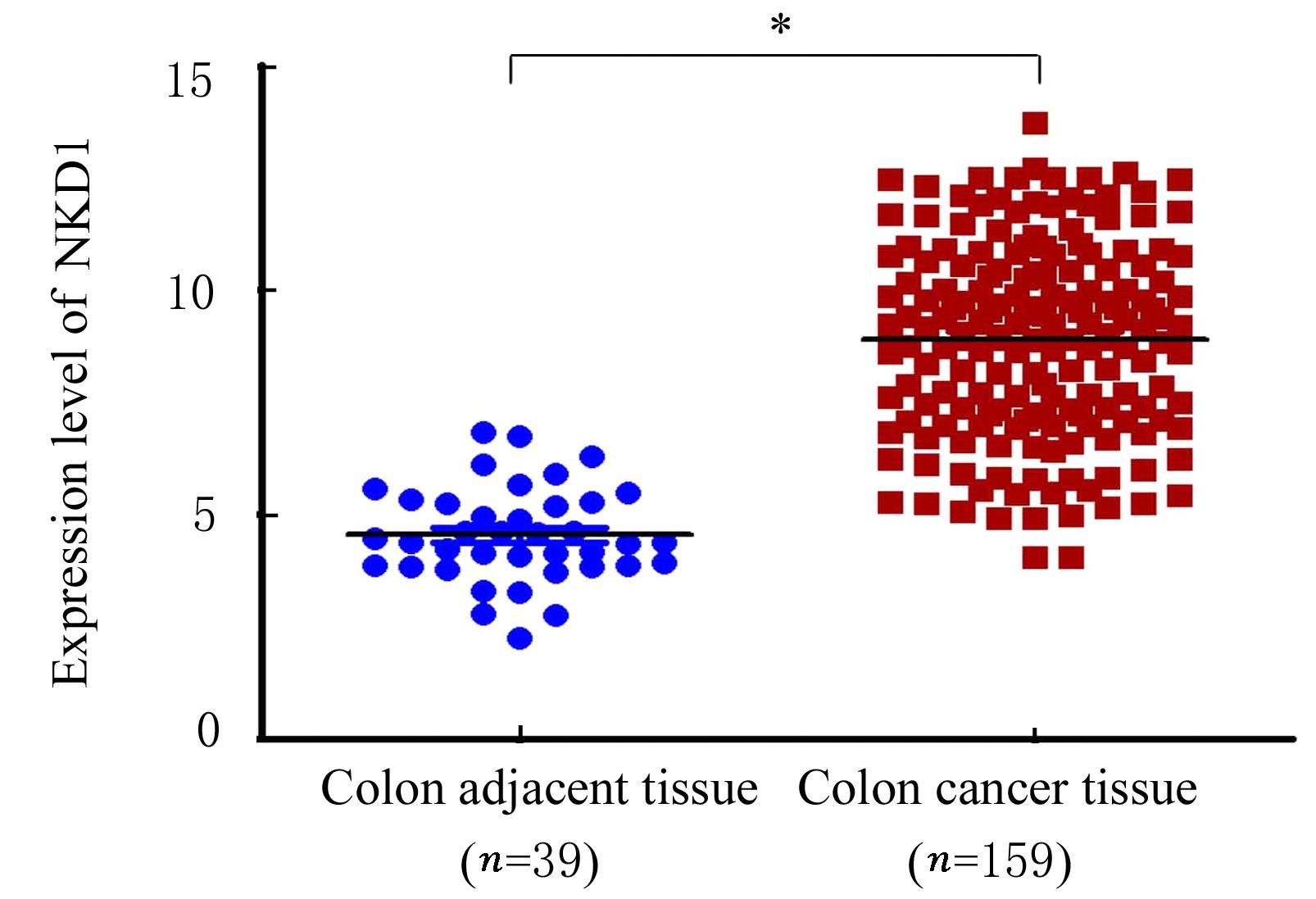
Journal of Jilin University(Medicine Edition) ›› 2022, Vol. 48 ›› Issue (3): 755-765.doi: 10.13481/j.1671-587X.20220325
• Research in clinical medicine • Previous Articles
Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors
Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN( )
)
- Department of Oncology,Affiliated Wujin Hospital,Jiangsu University,Wujin Clinical College,Xuzhou Medical University,Changzhou 213017,China
-
Received:2021-08-26Online:2022-05-28Published:2022-06-21 -
Contact:Jianhua JIN E-mail:jianhuajin88@sina.com
CLC Number:
- R735.35
Cite this article
Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors[J].Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765.
share this article
Tab. 1
Univariate and multivariate risk analysis on expression of AXIN2 gene and OS of colon cancer patients"
| Clinical parameter | Univariate risk analysis | Multivariate risk analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | |
| Age | 1.03 | 1.01-1.05 | 0.008 | 1.04 | 1.01-1.06 | 0.001 |
| Gender | 1.11 | 0.70-1.77 | 0.653 | 10.5 | 0.65-1.69 | 0.836 |
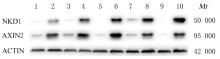
| Stage | 2.26 | 1.73-2.93 | 1.59E-09 | 1.21 | 0.57-2.60 | 0.622 |
| T | 2.85 | 1.80-4.52 | 7.77E-06 | 1.76 | 1.01-3.07 | 0.045 |
| M | 4.45 | 2.74-7.21 | 1.40E-09 | 2.28 | 0.81-6.41 | 0.118 |
| N | 2.02 | 1.54-2.65 | 3.47E-07 | 1.42 | 0.88-2.28 | 0.150 |
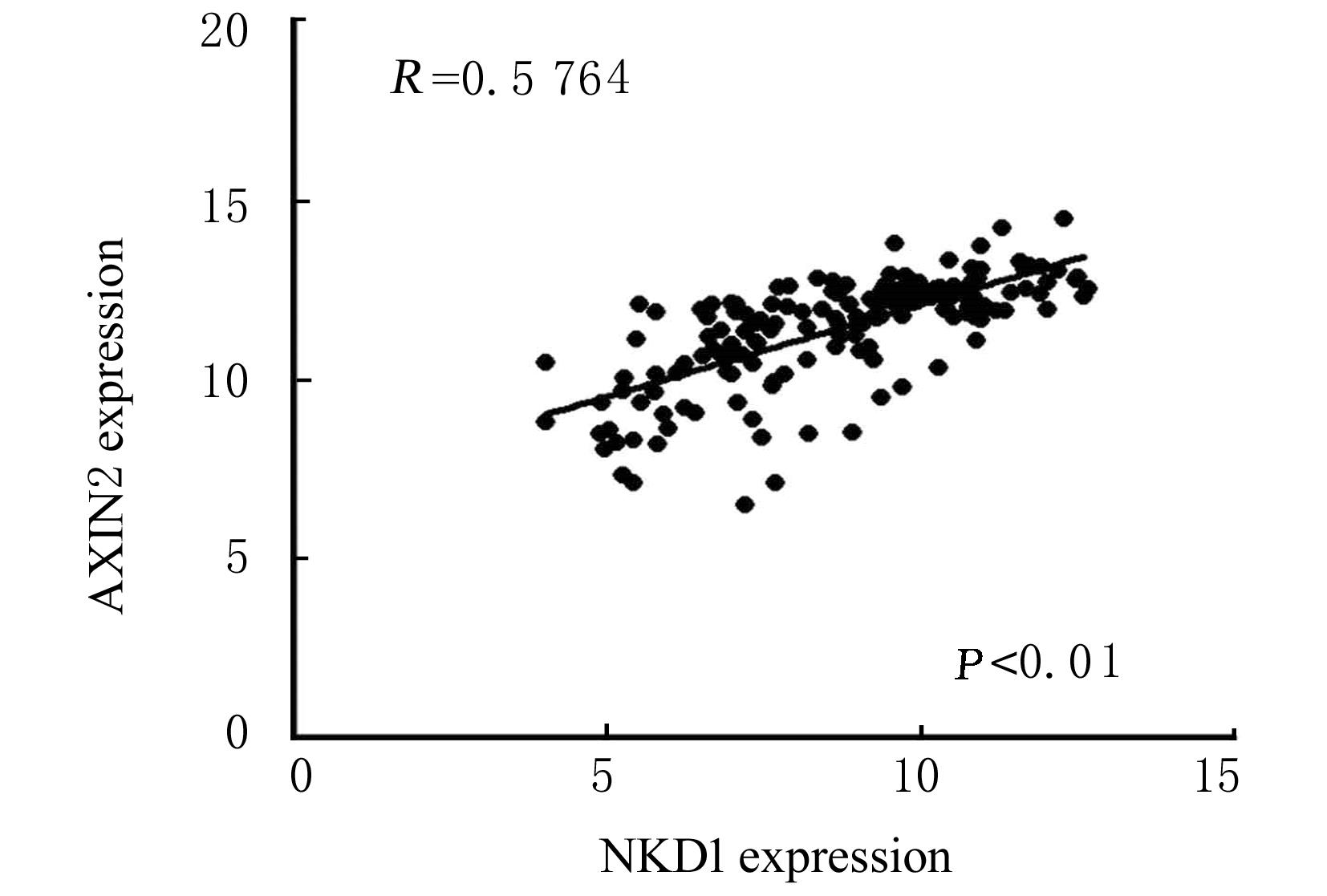
| AXIN2 | 0.99 | 0.98-1.00 | 0.023 | 0.82 | 0.69-0.98 | 0.026 |
| 1 | SIEGEL R L, MILLER K D, JEMAL A. Cancer statistics, 2020[J].CA Cancer J Clin,2020,70(1): 7-30. |
| 2 | FENG X D, ZHANG Z J, SUN P, et al. Interleukin-18 is a prognostic marker and plays a tumor suppressive role in colon cancer[J]. Dis Markers, 2020, 2020: 6439614. |
| 3 | ANGELESCU R, DOBRESCU R. MIDGET: Detecting differential gene expression on microarray data[J]. Comput Methods Programs Biomed,2021, 211: 106418. |
| 4 | WANG Y H, ZHAO Y, BOLLAS A, et al. Nanopore sequencing technology, bioinformatics and applications[J].Nat Biotechnol,2021,39(11):1348-1365. |
| 5 | CZECH L, HUERTA-CEPAS J, STAMATAKIS A. A critical review on the use of support values in tree viewers and bioinformatics toolkits[J]. Mol Biol Evol, 2017, 34(6): 1535-1542. |
| 6 | MA J Y, CHEN X C, LIN M Q, et al. Bioinformatics analysis combined with experiments predicts CENPK as a potential prognostic factor for lung adenocarcinoma[J]. Cancer Cell Int, 2021, 21(1): 65. |
| 7 | WANG Y C, TIAN Z B, TANG X Q. Bioinformatics screening of biomarkers related to liver cancer[J]. BMC Bioinformatics, 2021, 22(): 521. |
| 8 | 赵昕辉, 刘 俊, 贺奋飞, 等. 基于生物信息学分析的结肠癌枢纽基因筛选及调控网络构建[J]. 现代肿瘤医学, 2019, 27(13): 2227-2231. |
| 9 | DIBOUN I, WERNISCH L, ORENGO C A, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma[J]. BMC Genomics, 2006, 7: 252. |
| 10 | ROBINSON M D, MCCARTHY D J, SMYTH G K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data[J]. Bioinformatics, 2010, 26(1): 139-140. |
| 11 | ITO K, MURPHY D. Application of ggplot2 to pharmacometric graphics[J]. CPT Pharmacometrics Syst Pharmacol, 2013, 2(10): e79. |
| 12 | LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559. |
| 13 | YOUNG A, WHITEHOUSE N, CHO J, et al. OntologyTraverser: An R package for GO analysis[J]. Bioinformatics, 2005, 21(2): 275-276. |
| 14 | KANEHISA M, GOTO S, FURUMICHI M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs[J]. Nucleic Acids Res, 2010, 38(Database issue): D355-D360. |
| 15 | STEENWYK J L, ROKAS A. Ggpubfigs: colorblind-friendly color palettes and ggplot2 graphic system extensions for publication-quality scientific figures[J]. Microbiol Resour Announc, 2021, 10(44): e0087121. |
| 16 | ZHOU L N, DING L Y, GONG Y Q, et al. Identification of hub genes associated with the pathogenesis of diffuse large B-cell lymphoma subtype one characterized by host response via integrated bioinformatic analyses[J]. Peer J, 2020, 8: e10269. |
| 17 | LIU Z C, LIU X L, CAI R, et al. Identification of a tumor microenvironment-associated prognostic gene signature in bladder cancer by integrated bioinformatic analysis[J].Int J Clin Exp Pathol,2021,14(5): 551-566. |
| 18 | WANG Y, LUO H, CAO J, et al. Bioinformatic identification of neuroblastoma microenvironment-associated biomarkers with prognostic value[J].J Oncol, 2020, 2020: 5943014. |
| 19 | LIU Q, LU W B, YANG C X, et al. HBXIP activates the PPARδ/NF-κB feedback loop resulting in cell proliferation[J]. Oncotarget, 2018, 9(1): 404-417. |
| 20 | LIU Q, DENG J Z, YANG C X, et al. DPEP1 promotes the proliferation of colon cancer cells via the DPEP1/MYC feedback loop regulation[J]. Biochem Biophys Res Commun, 2020, 532(4): 520-527. |
| 21 | WANG Y, YANG C X, LI W J, et al. Identification of colon tumor marker NKD1 via integrated bioinformatics analysis and experimental validation[J]. Cancer Med, 2021, 10(20): 7383-7394. |
| 22 | STANCIKOVA J, KRAUSOVA M, KOLAR M, et al. NKD1 marks intestinal and liver tumors linked to aberrant Wnt signaling[J]. Cell Signal, 2015, 27(2): 245-256. |
| 23 | CAO B P, YANG W L, JIN Y S, et al. Silencing NKD2 by promoter region hypermethylation promotes esophageal cancer progression by activating Wnt signaling[J]. J Thorac Oncol, 2016, 11(11): 1912-1926. |
| 24 | BERNKOPF D B, BRÜCKNER M, HADJIHANNAS M V,et al. An aggregon in conductin/axin2 regulates Wnt/β-catenin signaling and holds potential for cancer therapy[J]. Nat Commun, 2019, 10(1): 4251. |
| 25 | KING D J, FREIMANIS G, LASECKA-DYKES L, et al. A systematic evaluation of high-throughput sequencing approaches to identify low-frequency single nucleotide variants in viral populations[J]. Viruses, 2020, 12(10): 1187. |
| 26 | LIU H Y, ZHANG C J. Identification of differentially expressed genes and their upstream regulators in colorectal cancer[J]. Cancer Gene Ther, 2017, 24(6): 244-250. |
| 27 | HESARI A, RAJAB S, REZAEI M, et al. Knockdown of Sal-like 4 expression by siRNA induces apoptosis in colorectal cancer[J]. J Cell Biochem, 2019: 2019Feb16. |
| 28 | GUO J H, CAGATAY T, ZHOU G J, et al. Mutations in the human naked cuticle homolog NKD1 found in colorectal cancer alter Wnt/Dvl/beta-catenin signaling[J]. PLoS One, 2009, 4(11): e7982. |
| 29 | CHEN H Y, LANG Y D, LIN H N, et al. miR-103/107 prolong Wnt/β-catenin signaling and colorectal cancer stemness by targeting Axin2[J]. Sci Rep, 2019, 9: 9687. |
| 30 | TIENG F Y F, ABU N, SUKOR S, et al. L1CAM, CA9, KLK6, HPN, and ALDH1A1 as potential serum markers in primary and metastatic colorectal cancer screening[J]. Diagnostics (Basel), 2020, 10(7): 444. |
| 31 | CUBAS R, ZHANG S, LI M, et al. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway[J]. Mol Cancer, 2010, 9: 253. |
| 32 | VELOUDIS G, PAPPAS A, GOURGIOTIS S, et al. Assessing the clinical utility of Wnt pathway markers in colorectal cancer[J]. J BUON, 2017, 22(2): 431-436. |
| 33 | OTERO L, LACUNZA E, VASQUEZ V, et al. Variations in AXIN2 predict risk and prognosis of colorectal cancer[J]. B D J Open, 2019, 5: 13. |
| [1] | Suxian CHEN,Zehui GU,Yangfei MA,Qi TAN,Qi LI,Yadi WANG. Promotion effect of rutin on apoptosis of human colon cancer SW480 cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 356-363. |
| [2] | Xiaoyan LI,Wei ZHANG,Jie HE. Promotion effect of REG1A on proliferation and migration of lung adenocarcinoma cells by regulating Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 444-453. |
| [3] | Ying ZHAO,Danyu ZHAO,Chao LIU. Bioinformatics analysis on prognostic evaluation value of TXNDC11 gene in pan-cancer and its immunity regulation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 142-153. |
| [4] | Zhijuan ZHAO,Lian MENG,Chunxia LIU. Bioinformatics analysis on miRNA-mRNA regulatory networks based on fusion genes acting in rhabdomyosarcoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 154-162. |
| [5] | Nan LI,Lei CHEN,Tianmin XU,Kun ZHANG. Bioinformatics analysis on screening of extracellular matrix related genes in patients with endometriosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 188-194. |
| [6] | Hao DONG,Haizhen JIANG,Chao LI,Dengke GAO,Yaping JIN,Huatao CHEN. Construction of mouse NR1D1 gene overexpression vector and its bioinformatics analysis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 94-103. |
| [7] | Yang YU,Sainan LIU,Yunkai LIU,Yong LI,Yichun QIAO,Yi CHENG. Bioinformatic analysis on expression characteristics of PRPS2 and its relationship with prognosis of breast cancer [J]. Journal of Jilin University(Medicine Edition), 2021, 47(5): 1229-1236. |
| [8] | Bo MA, Jiangang LI, Jun WANG, Junli HOU, Liang LI. Promotion effect of miR-106b on invasion and migration of colon cancer cells through targeting TGF-β/Smad pathway [J]. Journal of Jilin University(Medicine Edition), 2021, 47(3): 630-636. |
| [9] | Yuqing PAN, Yunyan SUN, Yixun LI, Liying WANG, Zhuoma SINAN, Rui CHEN, Xi ZHANG, Yan DU. Construction and analysis of competitive endogenous RNA networks in acute myeloid leukemia based on high-throughput microarray [J]. Journal of Jilin University(Medicine Edition), 2021, 47(3): 669-676. |
| [10] | Yao LIN,Chunlin LIN,Qin WANG,Bin ZHANG,Shaoyu WANG,Guangwei ZHU. Expressions of minichromosome maintenance proteins and structural maintenance of chromosomes 4 genes in cervical squamous cell carcinoma tissue and bioinformation analysis [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 430-437. |
| [11] | Lan ZHENG,Songzhe PIAO,Ran XU,Xinyue WANG,Yixuan WANG,Zhenhua LIN,Yang YANG. Bioinformatics analysis based on effect of expression levels of m6A regulators on immune infiltration and prognosis of uterine corpus endometrial carcinoma [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 438-452. |
| [12] | Weili HUANG,Yongchen LYU. Expressions of miR-1915-3p and Bcl-2 in human colon cancer and their clinical significances [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 453-459. |
| [13] | ZHANG Xi, ZHAI Li, SUN Yunyan, YANG Wei, GAO Yanzhang, LEI Ming, PAN Yuqing. Bioinformatics analysis of pediatric acute myeloid leukemia based on high-throughput microarray [J]. Journal of Jilin University(Medicine Edition), 2020, 46(05): 1036-1042. |
| [14] | WANG Zhi, HONG Li, LI Suting, ZENG Wanling. Analysis on endometrial cancer-related genes and candidate pathways based on GEO database bioinformatics methods [J]. Journal of Jilin University(Medicine Edition), 2020, 46(04): 804-809. |
| [15] | YAN Shengyu, XIE Yafeng, XU Zhijie, LIU Ying, DING Yating, ZHANG Qiao, LIU Wanying, LIU Libing. Inhibitory effect of antimicrobial peptide LL-37 on tumorgrowth of mice with colon cancer and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2020, 46(03): 575-581. |
|
||