Journal of Jilin University(Medicine Edition) ›› 2024, Vol. 50 ›› Issue (6): 1664-1676.doi: 10.13481/j.1671-587X.20240620
• Research in clinical medicine • Previous Articles
Bioinformatics analysis on PDE1B expression and prognosis of gastric cancer and tumor microenvironment
Xi YANG1,Qin YUAN2,Lan YANG1,3,Wenjie ZHANG1( )
)
- 1.Department of Pathology,School of Medical Sciences,Shihezi Sciences University,Shihezi 832002,China
2.Department of Oncology,Hubei Aerospace Hospital,Xiaogan 432000,China
3.Department of Pathology,First Affiliated Hospital,Shihezi University,Shihezi 832002,China
-
Received:2023-12-06Online:2024-11-28Published:2024-12-10 -
Contact:Wenjie ZHANG E-mail:1939453950@qq.com
CLC Number:
- R735.2
Cite this article
Xi YANG,Qin YUAN,Lan YANG,Wenjie ZHANG. Bioinformatics analysis on PDE1B expression and prognosis of gastric cancer and tumor microenvironment[J].Journal of Jilin University(Medicine Edition), 2024, 50(6): 1664-1676.
share this article
Tab.1
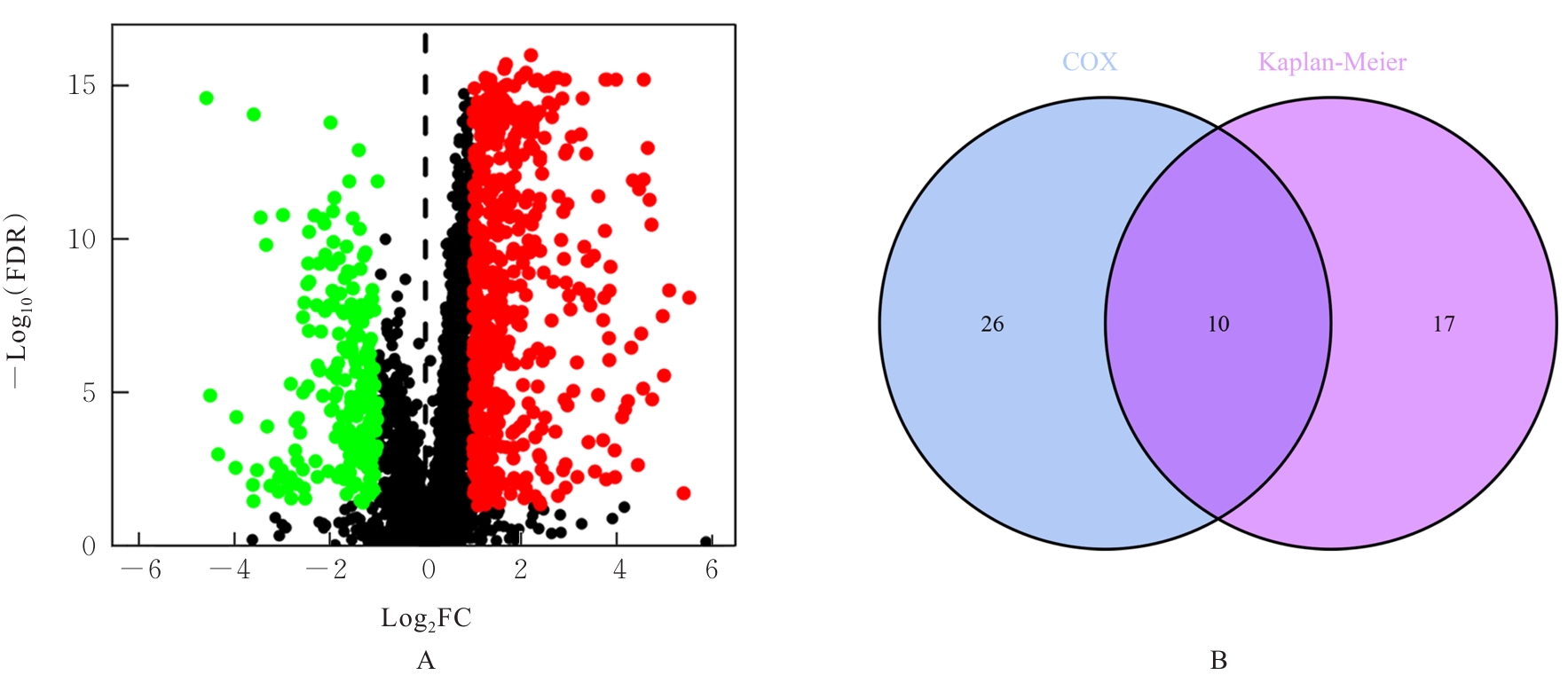
Relationships between intersecting genes and clinicopathological characteristics of gastric cancer patients"
| ID | P value of gender | P value of age | P value of grade | P value of stage | P value of TNM | Significative number | ||
|---|---|---|---|---|---|---|---|---|
| T | N | M | ||||||
| PDE1B | 0.670 0 | 0.027 0 | 6.8E-07 | 0.002 6 | 0.008 2 | 0.007 6 | 0.042 0 | 6 |
| AKAP12 | 0.390 0 | 0.047 0 | 4.4E-05 | 0.027 0 | 1.7E-05 | 0.040 0 | 0.650 0 | 5 |
| GHR | 0.350 0 | 0.001 3 | 4.1E-06 | 0.002 4 | 0.000 2 | 0.290 0 | 0.380 0 | 4 |
| GGT5 | 0.480 0 | 0.022 0 | 6.4E-08 | 0.009 2 | 1.4E-07 | 0.130 0 | 0.530 0 | 4 |
| CDO1 | 0.920 0 | 0.001 8 | 9.4E-08 | 0.010 0 | 1.2E-06 | 0.230 0 | 0.280 0 | 4 |
| LOX | 0.520 0 | 0.280 0 | 0.000 3 | 0.013 0 | 7.4E-07 | 0.390 0 | 0.290 0 | 3 |
| PDGFRB | 0.280 0 | 0.850 0 | 3.4E-05 | 0.004 1 | 1.8E-06 | 0.260 0 | 0.620 0 | 3 |
| VCAN | 0.750 0 | 0.800 0 | 0.000 5 | 0.018 0 | 2.1E-07 | 0.200 0 | 0.780 0 | 3 |
| TMEM200A | 0.078 0 | 0.100 0 | 0.230 0 | 0.017 0 | 0.002 0 | 0.240 0 | 0.820 0 | 2 |
| SERPINE1 | 0.850 0 | 0.410 0 | 0.022 0 | 0.140 0 | 0.003 4 | 0.430 0 | 0.520 0 | 2 |
| 1 | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| 2 | PARK E H, JUNG K W, PARK N J, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2021[J]. Cancer Res Treat, 2024, 56(2): 357-371. |
| 3 | ITO Y, MIYASHIRO I, ISHIKAWA T, et al. Determinant factors on differences in survival for gastric cancer between the United States and Japan using nationwide databases[J]. J Epidemiol, 2021, 31(4): 241-248. |
| 4 | GALLUZZI L, VITALE I, AARONSON S A, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018[J]. Cell Death Differ, 2018, 25(3): 486-541. |
| 5 | SCHMAUCK-MEDINA T, MOLIÈRE A, LAUTRUP S, et al. New hallmarks of ageing: a 2022 Copenhagen ageing meeting summary[J]. Aging, 2022, 14(16): 6829-6839. |
| 6 | COLLADO M, BLASCO M A, SERRANO M. Cellular senescence in cancer and aging[J]. Cell, 2007, 130(2): 223-233. |
| 7 | ZHUO S H, CHEN Z M, YANG Y B, et al. Clinical and biological significances of a ferroptosis-related gene signature in glioma[J]. Front Oncol, 2020, 10: 590861. |
| 8 | AHLUWALIA P, AHLUWALIA M, MONDAL A K, et al. Immunogenomic gene signature of cell-death associated genes with prognostic implications in lung cancer[J]. Cancers, 2021, 13(1): 155. |
| 9 | OKI E, TOKUNAGA S, EMI Y, et al. Surgical treatment of liver metastasis of gastric cancer: a retrospective multicenter cohort study (KSCC1302)[J]. Gastric Cancer, 2016, 19(3): 968-976. |
| 10 | AL-BATRAN S E, HOZAEEL W, JÄGER E. Combination of trastuzumab and triple FLOT chemotherapy (5-fluorouracil/leucovorin, oxaliplatin, and docetaxel) in patients with HER2-positive metastatic gastric cancer: report of 3 cases[J]. Onkologie, 2012, 35(9): 505-508. |
| 11 | BANG Y J, VAN CUTSEM E, FEYEREISLOVA A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial[J]. Lancet, 2010, 376(9742): 687-697. |
| 12 | MITSUI Y, SATO Y, MIYAMOTO H, et al. Trastuzumab in combination with docetaxel/cisplatin/S-1 (DCS) for patients with HER2-positive metastatic gastric cancer: feasibility and preliminary efficacy[J]. Cancer Chemother Pharmacol, 2015, 76(2): 375-382. |
| 13 | ZHAO C G, MO L P, LEI T, et al. MiR-5701 promoted apoptosis of clear cell renal cell carcinoma cells by targeting phosphodiesterase-1B[J]. Anticancer Drugs, 2021, 32(8): 855-863. |
| 14 | CHEUNG W Y. Cyclic 3',5'-nucleotide phosphodiesterase[J]. Biochem Biophys Res Commun, 1970, 38(3): 533-538. |
| 15 | BOLGER G B. The cAMP-signaling cancers: clinically-divergent disorders with a common central pathway[J]. Front Endocrinol, 2022, 13: 1024423. |
| 16 | ZHANG H Y, KONG Q B, WANG J, et al. Complex roles of cAMP-PKA-CREB signaling in cancer[J]. Exp Hematol Oncol, 2020, 9(1): 32. |
| 17 | KUMAZOE M, TAKAI M K, HIROI S, et al. The FOXO3/PGC-1β signaling axis is essential for cancer stem cell properties of pancreatic ductal adenocarcinoma[J]. J Biol Chem, 2017, 292(26): 10813-10823. |
| 18 | KUMAZOE M, SUGIHARA K, TSUKAMOTO S, et al. 67-kDa laminin receptor increases cGMP to induce cancer-selective apoptosis[J]. J Clin Invest, 2013, 123(2): 787-799. |
| 19 | YAMASHITA M, KUMAZOE M, ONDA H, et al. PPAR/PDK4 pathway is involved in the anticancer effects of cGMP in pancreatic cancer[J]. Biochem Biophys Res Commun, 2023, 672: 154-160. |
| 20 | SONNENBURG W K, SEGER D, BEAVO J A. Molecular cloning of a cDNA encoding the “61-kDa” calmodulin-stimulated cyclic nucleotide phosphodiesterase. Tissue-specific expression of structurally related isoforms[J]. J Biol Chem, 1993, 268(1): 645-652. |
| 21 | BENDER A T, OSTENSON C L, GIORDANO D, et al. Differentiation of human monocytes in vitro with granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor produces distinct changes in cGMP phosphodiesterase expression[J]. Cell Signal, 2004, 16(3): 365-374. |
| 22 | CHOW A, SCHAD S, GREEN M D, et al. Tim-4+ cavity-resident macrophages impair anti-tumor CD8+ Tcell immunity[J]. Cancer Cell, 2021, 39(7): 973-988. |
| 23 | KRYCZEK I, LANGE A, MOTTRAM P, et al. CXCL12 and vascular endothelial growth factor synergistically induce neoangiogenesis in human ovarian cancers[J]. Cancer Res, 2005, 65(2): 465-472. |
| 24 | GUO Q, GAO B L, ZHANG X J, et al. CXCL12-CXCR4 axis promotes proliferation, migration, invasion, and metastasis of ovarian cancer[J]. Oncol Res, 2014, 22(5/6): 247-258. |
| 25 | ZHANG F, CUI J Y, GAO H F, et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition and cisplatin resistance in ovarian cancer via CXCL12/CXCR4 axis[J]. Future Oncol, 2020, 16(32): 2619-2633. |
| 26 | LUKER K E, LUKER G D. The CXCL12/CXCR4/ACKR3 signaling axis regulates PKM2 and glycolysis[J]. Cells, 2022, 11(11): 1775. |
| 27 | ROCCONI R P, STANBERY L, TANG M, et al. ENTPD1/CD39 as a predictive marker of treatment response to gemogenovatucel-T as maintenance therapy in newly diagnosed ovarian cancer[J]. Commun Med, 2022, 2: 106. |
| 28 | THELEN M, LECHNER A, WENNHOLD K, et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8+ T cells-letter[J]. Cancer Res, 2018, 78(17): 5173-5174. |
| 29 | GU J, NI X H, PAN X X, et al. Human CD39hi regulatory T cells present stronger stability and function under inflammatory conditions[J]. Cell Mol Immunol, 2017, 14(6): 521-528. |
| [1] | Haiyao PANG,Jun MENG. Research progress in relationship between 14-3-3ɛ protein and occurrence and development of tumor [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 572-578. |
| [2] | Xiaodong GAI,Ying ZHAO,Hefei WANG,Chengyuan HE,Xingxiang WANG,Chun LI. Regulatory effect of FOXP3 on chemosensitivity of non small-cell lung cancer A549 cells to doxorubicin and its mechnism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1161-1167. |
| [3] | Yifei SUN,Dinuo LI,Yubin WANG. Inhibitory effect of curcumin on proliferation and invasion of gastric cancer MGC-803 cells by down-regulating PI3K/Akt/mTOR signaling pathway protein expression [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 332-340. |
| [4] | Renyi YANG,Shuwang PENG,Yongheng WANG,Yuxuan DONG,Shanshan DUAN. Construction of ferroptosis prognostic risk model of thyroid cancer and bioinformatics analysis on its potential mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 402-413. |
| [5] | Dehong ZHANG,Mingzhu ZHENG,Jiaqiu LI,Zhong LU. Bioinformatics analysis based on expressions of MSR1 mRNA and protein in pan-cancer tissue and its significance [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 425-439. |
| [6] | Yun LIU,Linqi ZHU,Shihe SHAO. Effects of circular RNA hsa_circ_0009735 on epithelial mesenchymal transformation, cell cycle, and autophagy of gastric cancer cells [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1498-1509. |
| [7] | Junjie HOU,Xuguang MI,Xiaonan LI,Xiaonan LI,Ying YANG,Xiaodan LU,Yanqiu FANG,Ningyi JIN. Establishment and evaluation of prognostic model of lung adenocarcinoma based on lactate metabolism gene [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1546-1554. |
| [8] | Xianshun XIE,Wei WANG,Haibing JIANG. Effect of miR-431-3p on proliferation and apoptosis of gastric cancer cells and its mechanism of targeted regulation of CTDP1 gene expression [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1555-1565. |
| [9] | Qian ZHANG,Jing LI. Inhibitory effect of TLR4 gene overexpression on autophagy of gastric cancer cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1238-1246. |
| [10] | Guangsong XU,Haibing JIANG,Jing PAN,Guoqing LI. Inhibitory effects of betulinic acid on migration and invasion of gastric cancer MGC-803 cells and their mechanisms [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 122-128. |
| [11] | Lili QIN,Xiaobo MA,Tianye ZHAO,Xuerong TAO,Min ZHENG,Xueying WANG,Jiaxin YI,Yanhua WU,Jing JIANG. Effects of MMP-9 and TIMP-1 expressions on prognostic evaluation of gastric cancer patients after radical gastrectomy [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 163-171. |
| [12] | Runhong MU,Yijiu AI,Yupeng LI,Rui LIN,Siping YE,Fang MA,Xiao GUO. Expression of recombinant human IL-17A in gastric cancer tissue and its effects on proliferation, invasion, migration and apoptosis of gastric cancer BGC-823 cells [J]. Journal of Jilin University(Medicine Edition), 2021, 47(6): 1510-1517. |
| [13] | Jingjing GE,Hongxia XU,Lixia XIE,Kaili DU,Ming SANG,Xiaodong SUN. Establishment of human serous ovarian cancer cell culture system in vitro and its application in chemotherapeutic drug sensitivity detection [J]. Journal of Jilin University(Medicine Edition), 2021, 47(4): 999-1007. |
| [14] | Nuan WANG,Lijuan YANG,Juanjuan DAI,Aili WANG,Yan WU,Chengxia LIU. Promotion effects of MARCH1 on migration and invasion of human gastric cancer cells through PI3K/AKT signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 352-359. |
| [15] | Ruiying SUN,Yang LI,Xu ZHAO,Linlin QU,Wei XU,Yunli ZHANG. Detection of serum levels of gastric cancer-related cytokines and their clinical significances in diagnosis of gastric cancer [J]. Journal of Jilin University(Medicine Edition), 2020, 46(6): 1274-1282. |
|
||