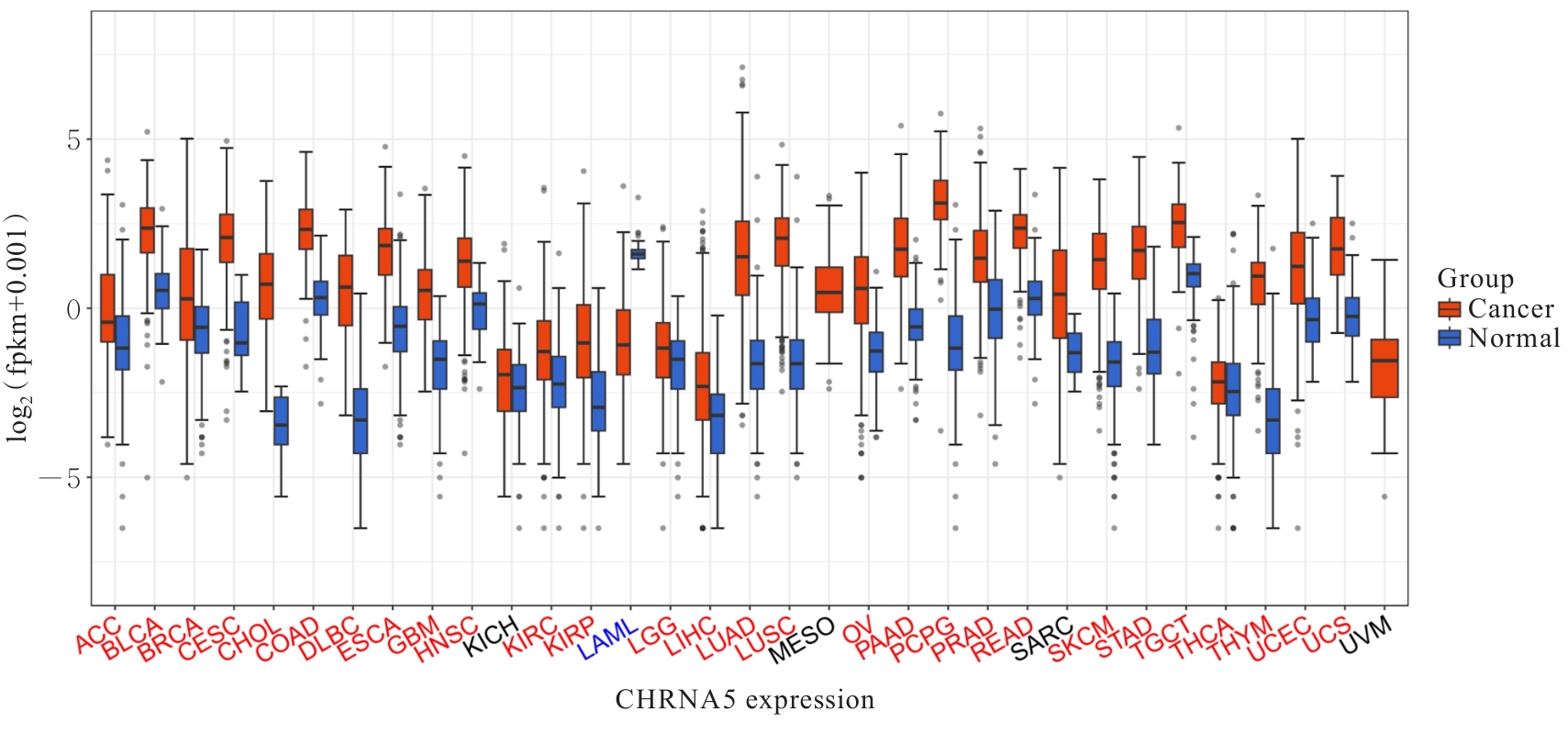
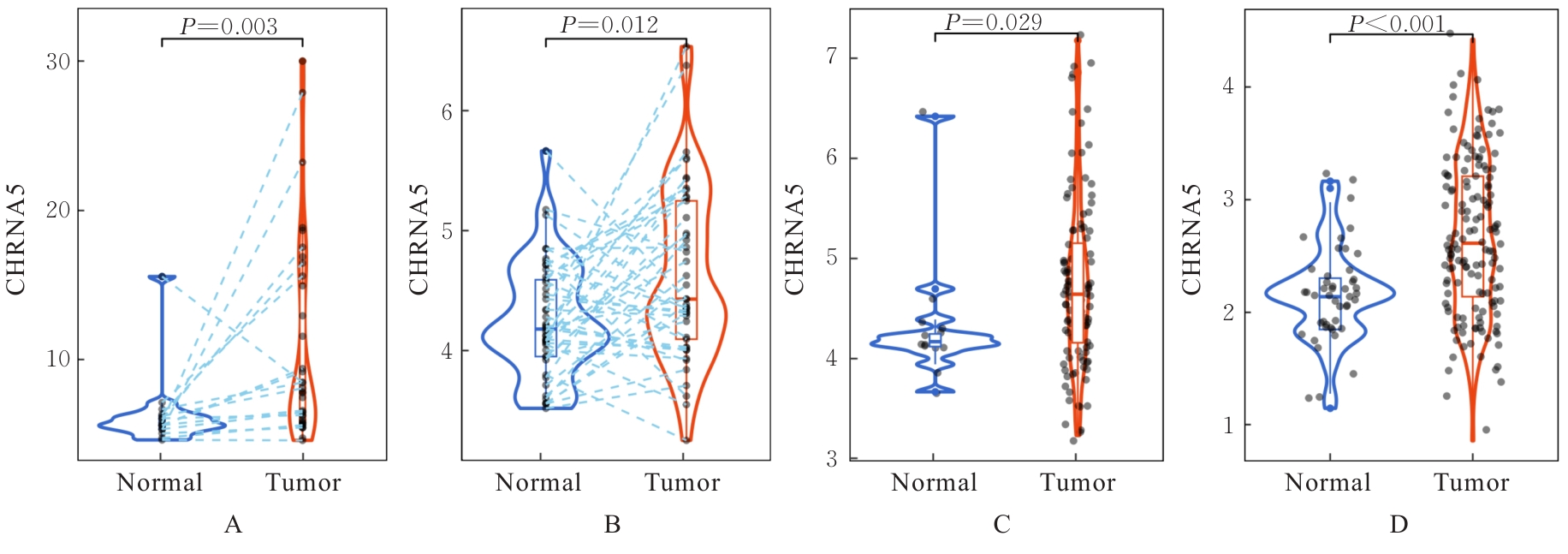
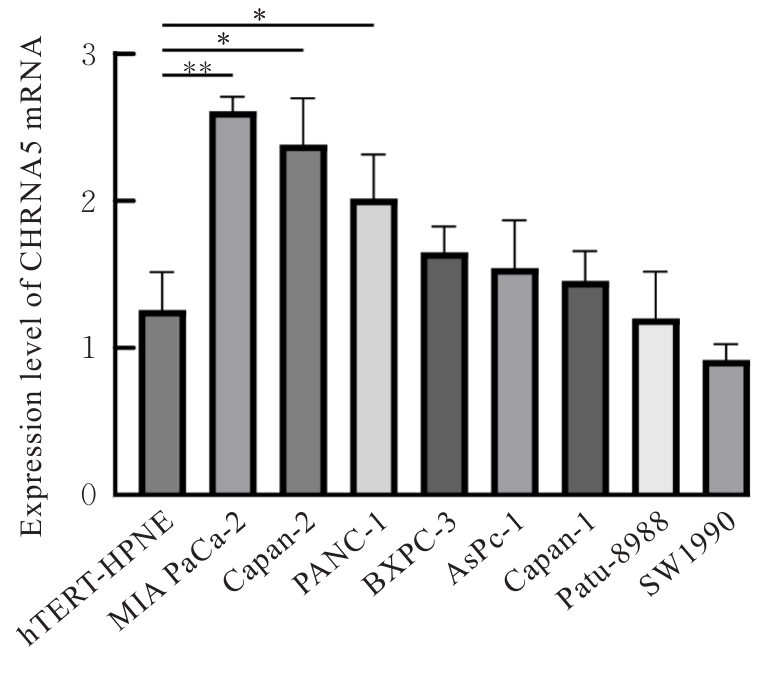
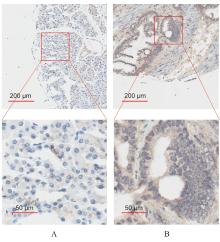
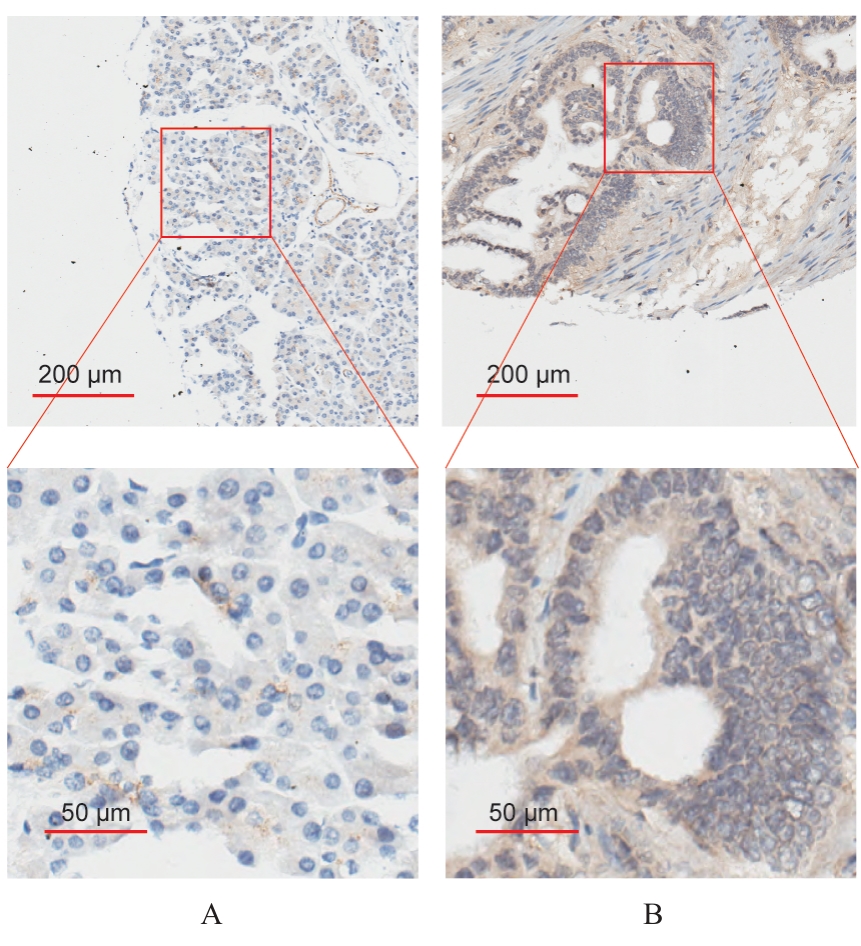
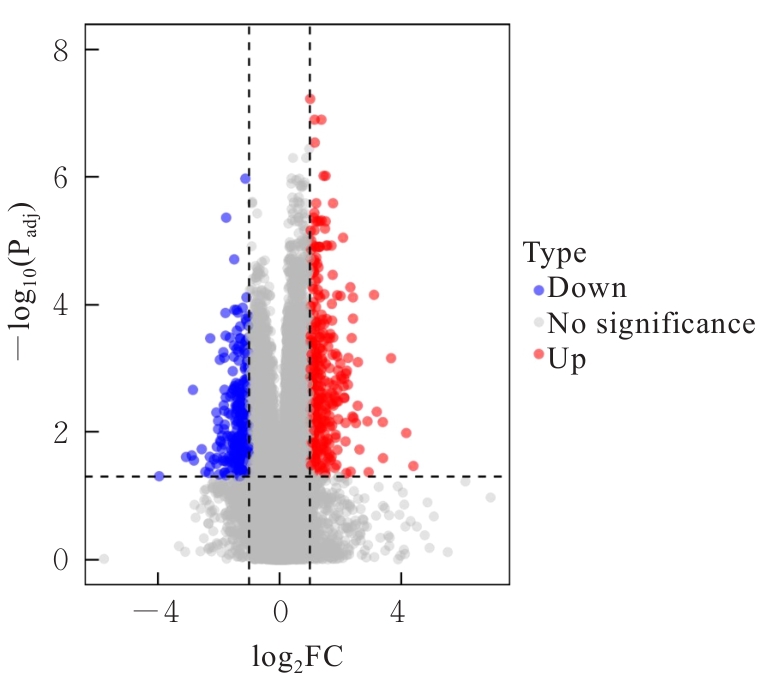
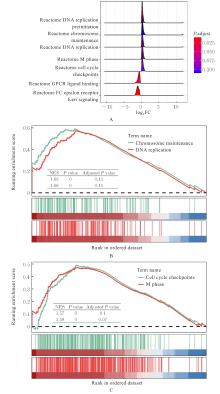
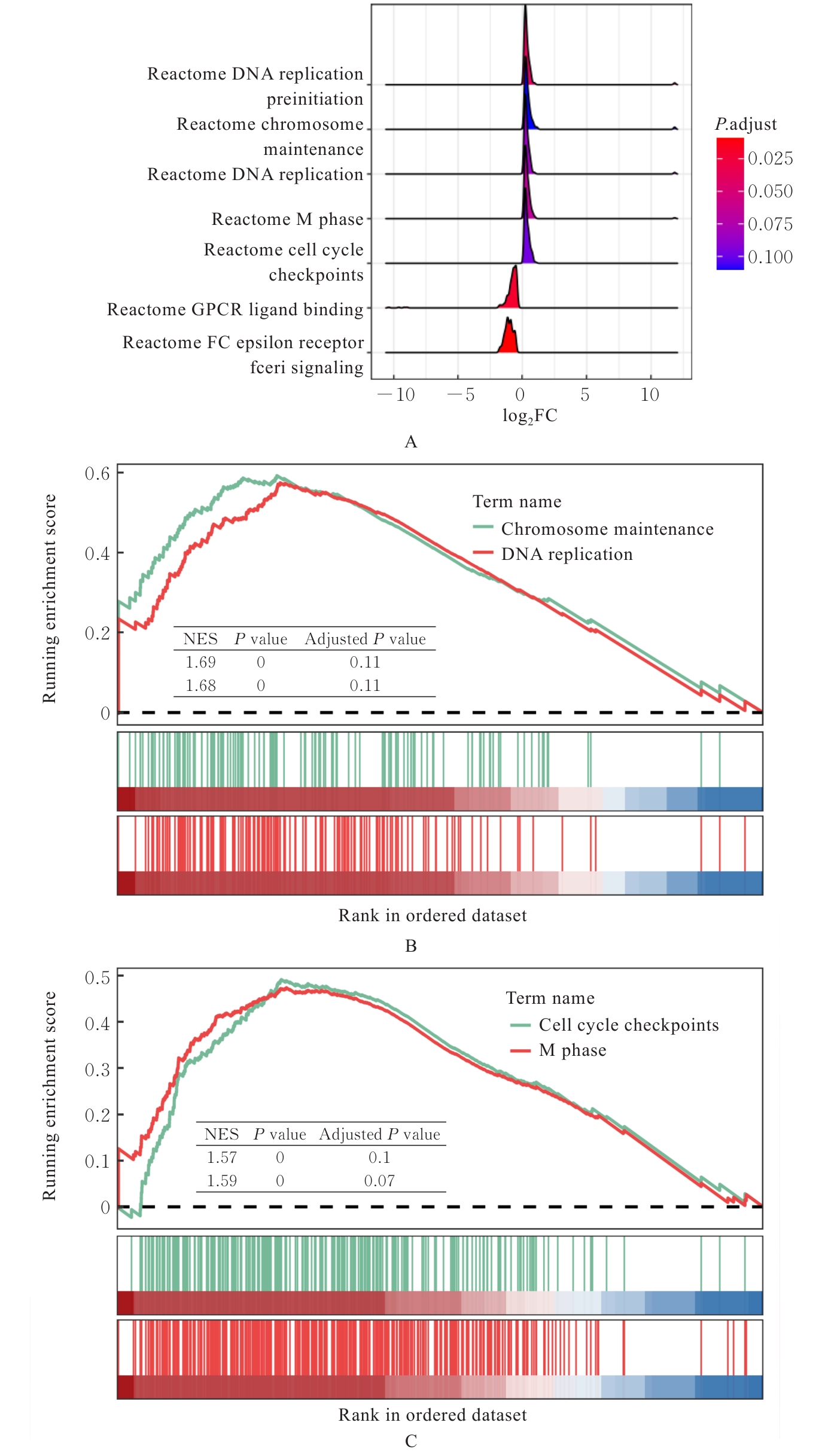
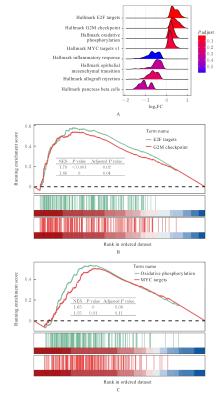
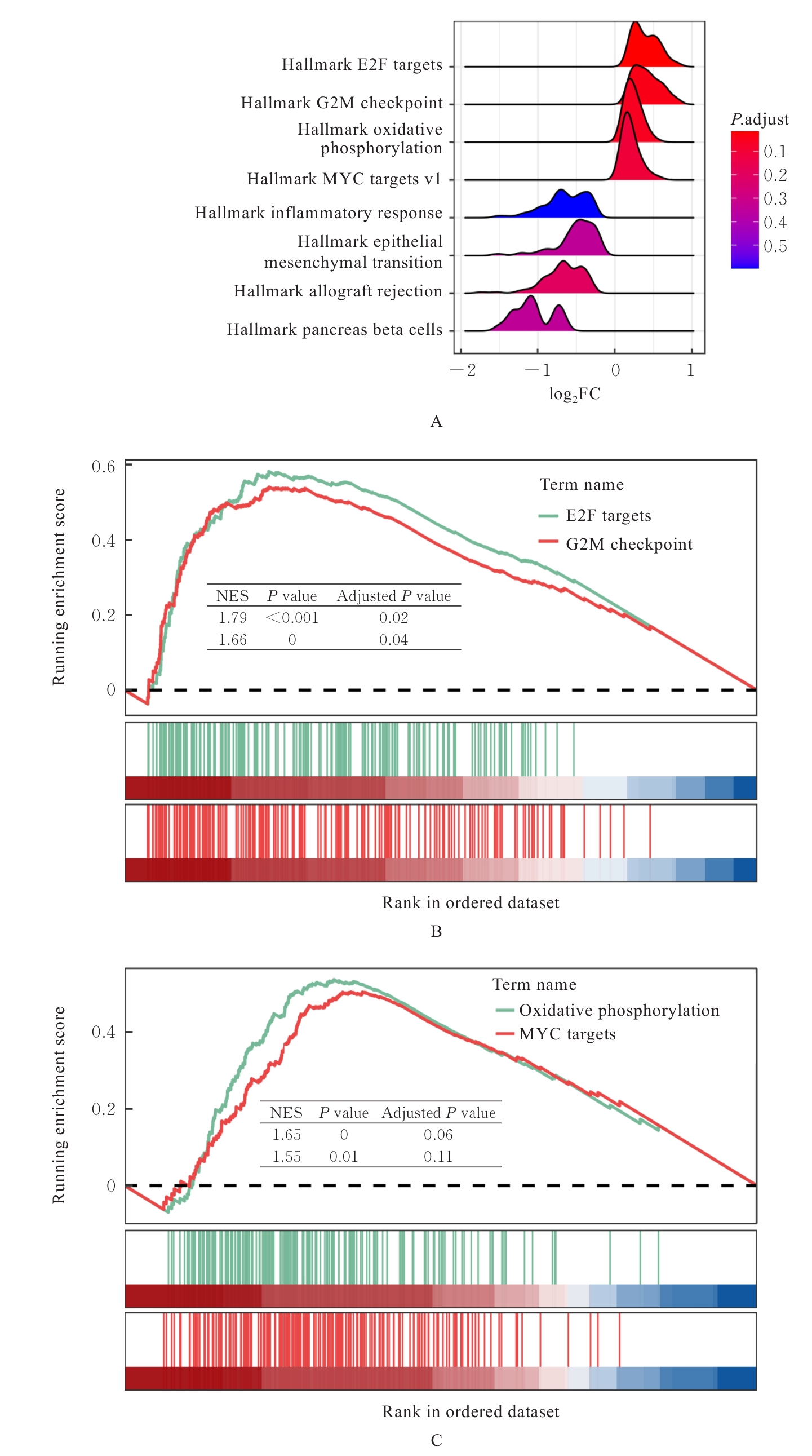
| 1 |
REBECCA L S, KIMBERLY D, NIKITA S W, et al. Cancer statistics, 2023[J] . CA Cancer J Clin, 2023, 73(1): 17-48.
|
| 2 |
MIZRAHI J D, SURANA R, VALLE J W, et al. Pancreatic cancer[J]. Lancet, 2020, 395(10242): 2008-2020.
|
| 3 |
SHERMAN M H, BEATTY G L,Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance[J] .Annu Rev Pathol, 2023, 18: 123-148.
|
| 4 |
CHY Y, MILLY M, ALBERT D, et al. Temporal changes in cause of death among adolescents and adults in six countries in eastern and southern Africa in 1995-2019: a multi-country surveillance study of verbal autopsy data[J]. Lancet Glob Health, 2024, 12(8): e1278-e1287.
|
| 5 |
JIANG S H, Hu L P, WANG Xet al. Neurotransmitters: emerging targets in cancer[J]. Oncogene, 2020, 39(3): 503-515.
|
| 6 |
LUTGENDORF S K, SOOD A K, ANTONI M H, Host factors and cancer progression : biobehavioral signaling pathways and interventions[J]. J Clin Oncol, 2010, 28(26): 4094-4099.
|
| 7 |
YANG Y H, LIU J B, GUI Y, et al. Relationship between autophagy and perineural invasion, clinicopathological features, and prognosis in pancreatic cancer[J]. World J Gastroenterol, 2017, 23(40): 7232-7241.
|
| 8 |
SALOMAN J L, ALBERS K M, LI D J, et al. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer[J]. Proc Natl Acad Sci U S A, 2016, 113(11): 3078-3083.
|
| 9 |
WANG X B, ROUZANNA I, YE L H, et al. Phenotype screens of murine pancreatic cancer identify a Tgf-α-Ccl2-paxillin axis driving human-like neural invasion[J]. J Clin Invest, 2023, 133(21): e166333.
|
| 10 |
KRAIS A M, HAUTEFEUILLE A H, CROS M P, et al. CHRNA5 as negative regulator of nicotine signaling in normal and cancer bronchial cells: effects on motility, migration and p63 expression[J]. Carcinogenesis, 2011, 32(9): 1388-1395.
|
| 11 |
WALTERS I B, BURACK L H, RCOVENet al T. Suberythemogenic narrow-band UVB is markedly more effective than conventional UVB in treatment of psoriasis vulgaris[J]. J Am Acad Dermatol, 1999, 40(6): 893-900.
|
| 12 |
MENTER A, KORMAN N J, ELMENTS C A, et al. Guidelines of care for the management of psoriasis and psoriatic arthritis: section 4. Guidelines of care for the management and treatment of psoriasis with traditional systemic agents[J]. J Am Acad Dermatol, 2009, 61(3): 451-85.
|
| 13 |
MENTER A, FELDMAN S R, WEINSTEIN G D, et al. A randomized comparison of continuous vs. intermittent infliximab maintenance regimens over 1 year in the treatment of moderate-to-severe plaque psoriasis[J]. J Am Acad Dermatol, 2007, 56(1): 31.e1-15.
|
| 14 |
KAUR-KNUDSEN D, BOJSEN S E, TYBJARG-HANSEN A, et al. Nicotinic acetylcholine receptor polymorphism, smoking behavior, and tobacco-related cancer and lung and cardiovascular diseases: a cohort study[J]. J Clin Oncol, 2011, 29(21): 2875-2882.
|
| 15 |
EMOKE R, DE LEEUW J, BAERVELDT E Met al. Cellular and molecular effects of pulsed dye laser and local narrow-band UVB therapy in psoriasis[J]. Lasers Surg Med, 2010, 42(3): 201-210.
|
| 16 |
ZOU H L, CHEN Y, ZHU X P, et al. Spinosad blocks CHRNA5 mediated EGFR signaling pathway activation to inhibit lung adenocarcinoma proliferation[J]. Biomed Pharmacother, 2024, 177: 117105.
|
| 17 |
FU Y, SHEN K Y, WANG H, et al. Alpha5 nicotine acetylcholine receptor subunit promotes intrahepatic cholangiocarcinoma metastasis[J]. Signal Transduct Target Ther, 2024, 9(1): 63.
|
| 18 |
ZAHALKA A H, FRENETTE P S, Nerves in cancer [J]. Nat Rev Cancer, 2020, 20(3): 143-157.
|
| 19 |
ZHANG Y, JIA Y F, LI P, et al. Reciprocal activation of α5-nAChR and STAT3 in nicotine-induced human lung cancer cell proliferation[J]. J Genet Genomics, 2017, 44(7): 355-362.
|
| 20 |
RUFFELL B, AFFARA N I, Coussens Lisa M, Differential macrophage programming in the tumor microenvironment[J]. Trends Immunol, 2012, 33(3): 119-126.
|
| 21 |
ZHANG W R, WANG M M, JI C H, et al. Macrophage polarization in the tumor microenvironment: Emerging roles and therapeutic potentials[J]. Biomed Pharmacother, 2024, 177: 116930.
|
| 22 |
LAUMONT C M, BANVILLE A C, GILARDI M, et al. Tumour-infiltrating B cells: immunological mechanisms, clinical impact and therapeutic opportunities[J]. Nat Rev Cancer, 2022, 22(7): 414-430.
|
| 23 |
JIN L, Kim H S, SHI J Q, Neutrophil in the Pancreatic Tumor Microenvironment [J]. Biomolecules, 2021, 11(8): 1170.
|
| 24 |
SILVESTRE-ROIG C, KALAFATI L, CHAVAKIS T, Neutrophils are shaped by the tumor microenvironment : novel possibilities for targeting neutrophils in cancer[J]. Signal Transduct Target Ther, 2024, 9(1): 77.
|
| 25 |
RENUMATHY D, ANJA, MAHAUAD-FERNANDEZ W D, et al. The MYC oncogene-the grand orchestrator of cancer growth and immune evasion [J]. Nat Rev Clin Oncol, 2022, 19(1): 23-36.
|
| 26 |
ARIANNA G, SARA T, GIORGIA G, et al. The FGF/FGFR/c-Myc axis as a promising therapeutic target in multiple myeloma [J]. J Exp Clin Cancer Res, 2024, 43(1): 294.
|
| 27 |
MCINTYRE C A, ADRIEN G, JIWOON P, et al. Distinct clinical outcomes and biological features of specific KRAS mutants in human pancreatic cancer [J]. Cancer Cell, 2024, 42(9): 1614-1629.e5.
|
| 28 |
MASUGI Y, TAKAMATSU M, TANAKA M, et al. Post-operative mortality and recurrence patterns in pancreatic cancer according to KRAS mutation and CDKN2A, p53, and SMAD4 expression[J]. J Pathol Clin Res, 2023, 9(5): 339-353.
|
 ),Hui LI(
),Hui LI( )
)