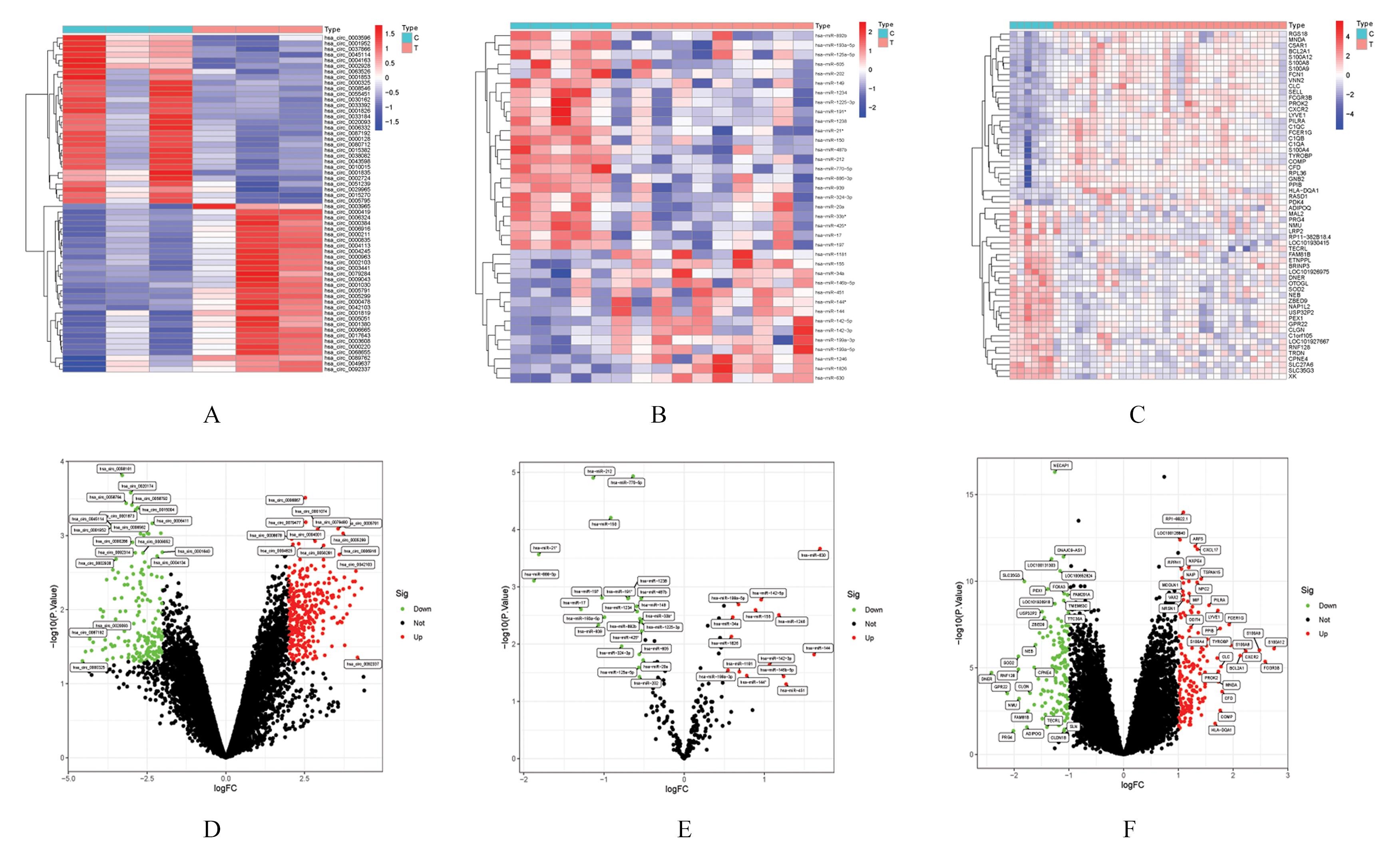
Journal of Jilin University(Medicine Edition) ›› 2022, Vol. 48 ›› Issue (6): 1535-1545.doi: 10.13481/j.1671-587X.20220620
• Research in clinical medicine • Previous Articles Next Articles
Bioinformatics analysis based on circRNA-miRNA-mRNA network construction and immune cell infiltration in atrial fibrillation
Jilin FAN1,Tingting ZHU2,Xiaoling TIAN2,Sijia LIU3,Jing SU3,Shiliang ZHANG3( )
)
- 1.First College of Clinical Medical Sciences,Shandong University of Traditional Chinese Medicine,Jinan 250014,China
2.Department of Neurosurgery,Affiliated Hospital,Binzhou Medical College,Binzhou 256600,China
3.Department of Cardiology,Affiliated Hospital,Shandong University of Traditional Chinese Medicine,Jinan 250014,China
-
Received:2021-11-23Online:2022-11-28Published:2022-12-07 -
Contact:Shiliang ZHANG E-mail:zhangshiliangsd@126.com
CLC Number:
- R541.75
Cite this article
Jilin FAN,Tingting ZHU,Xiaoling TIAN,Sijia LIU,Jing SU,Shiliang ZHANG. Bioinformatics analysis based on circRNA-miRNA-mRNA network construction and immune cell infiltration in atrial fibrillation[J].Journal of Jilin University(Medicine Edition), 2022, 48(6): 1535-1545.
share this article
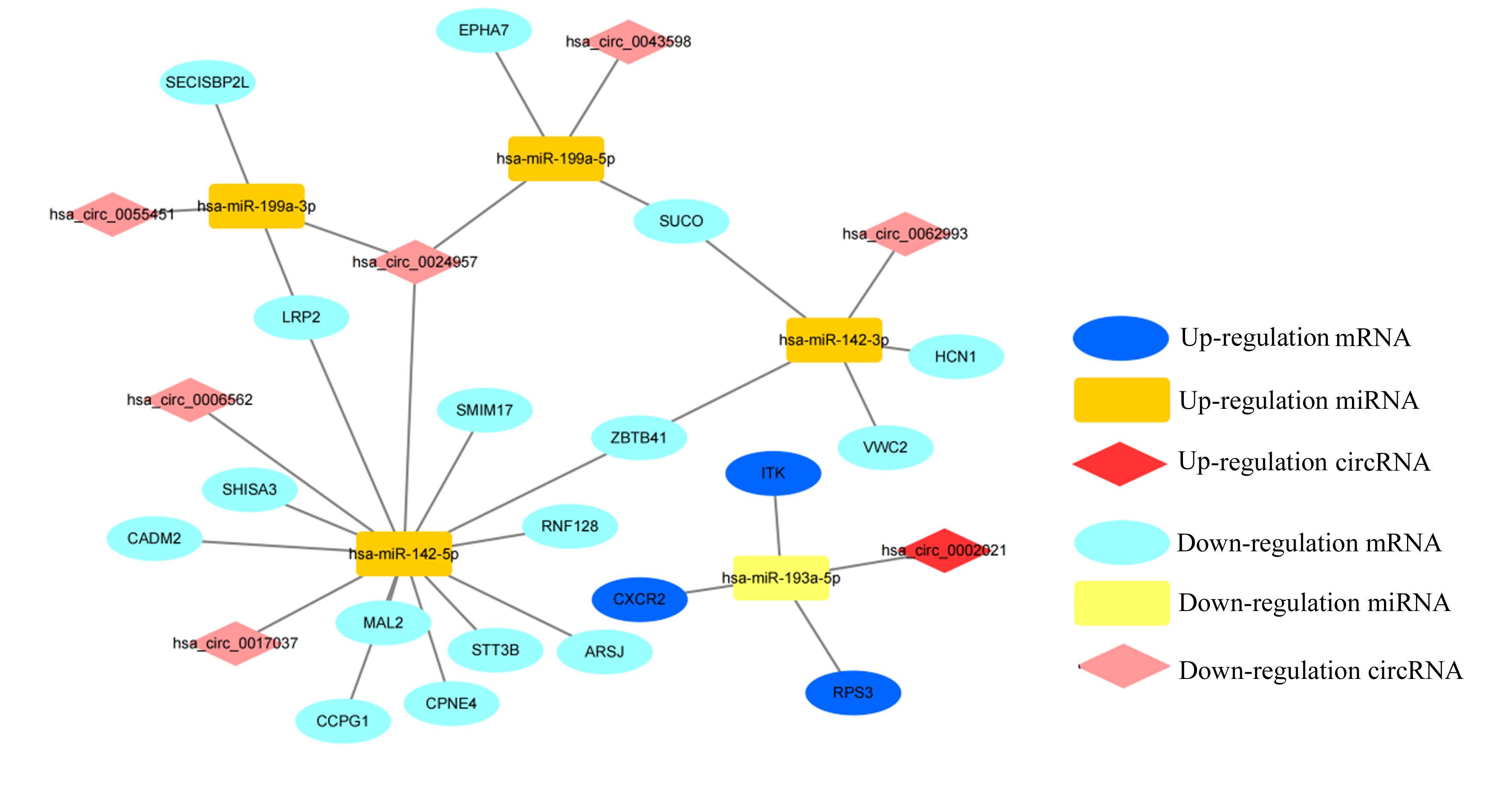
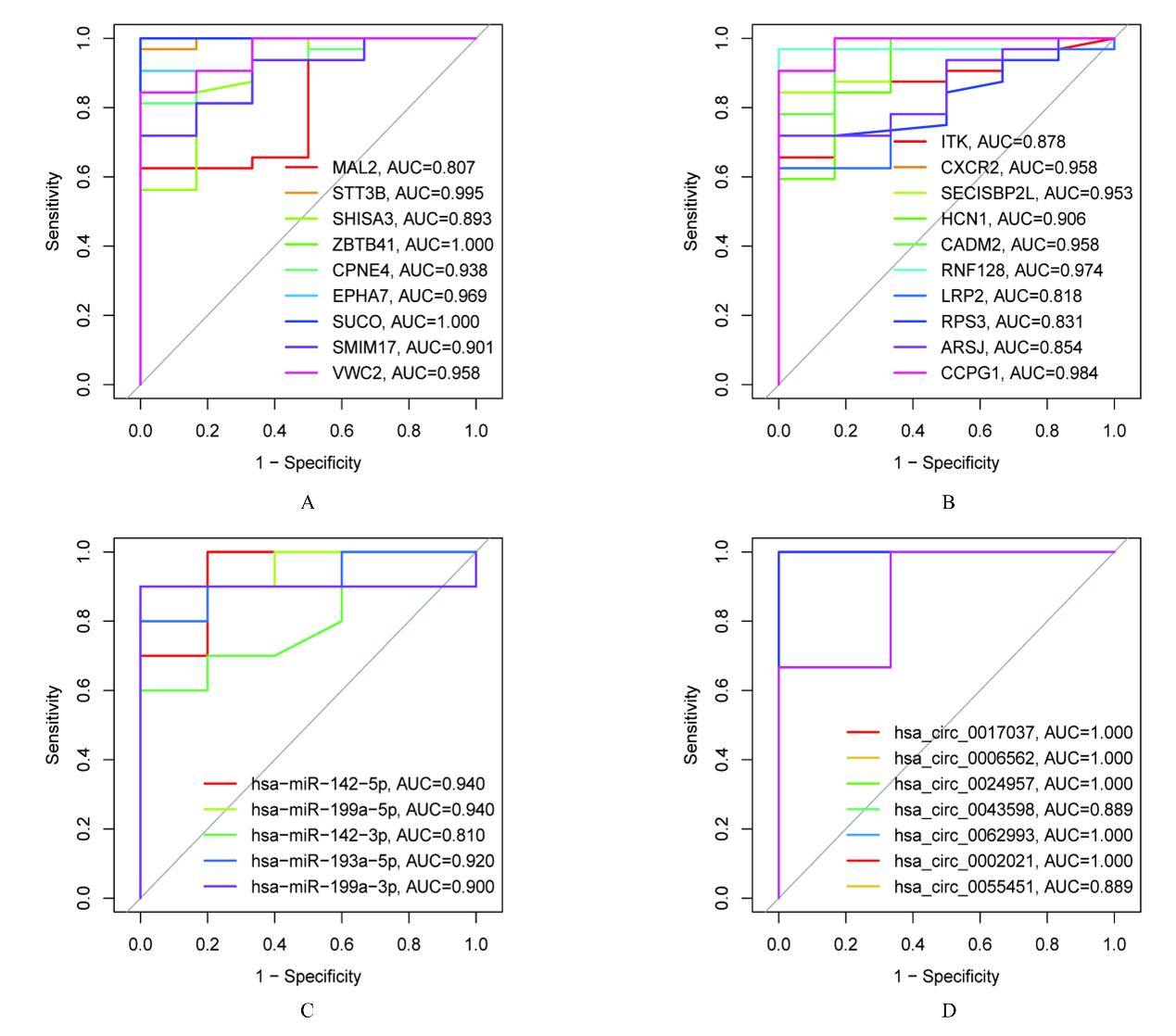
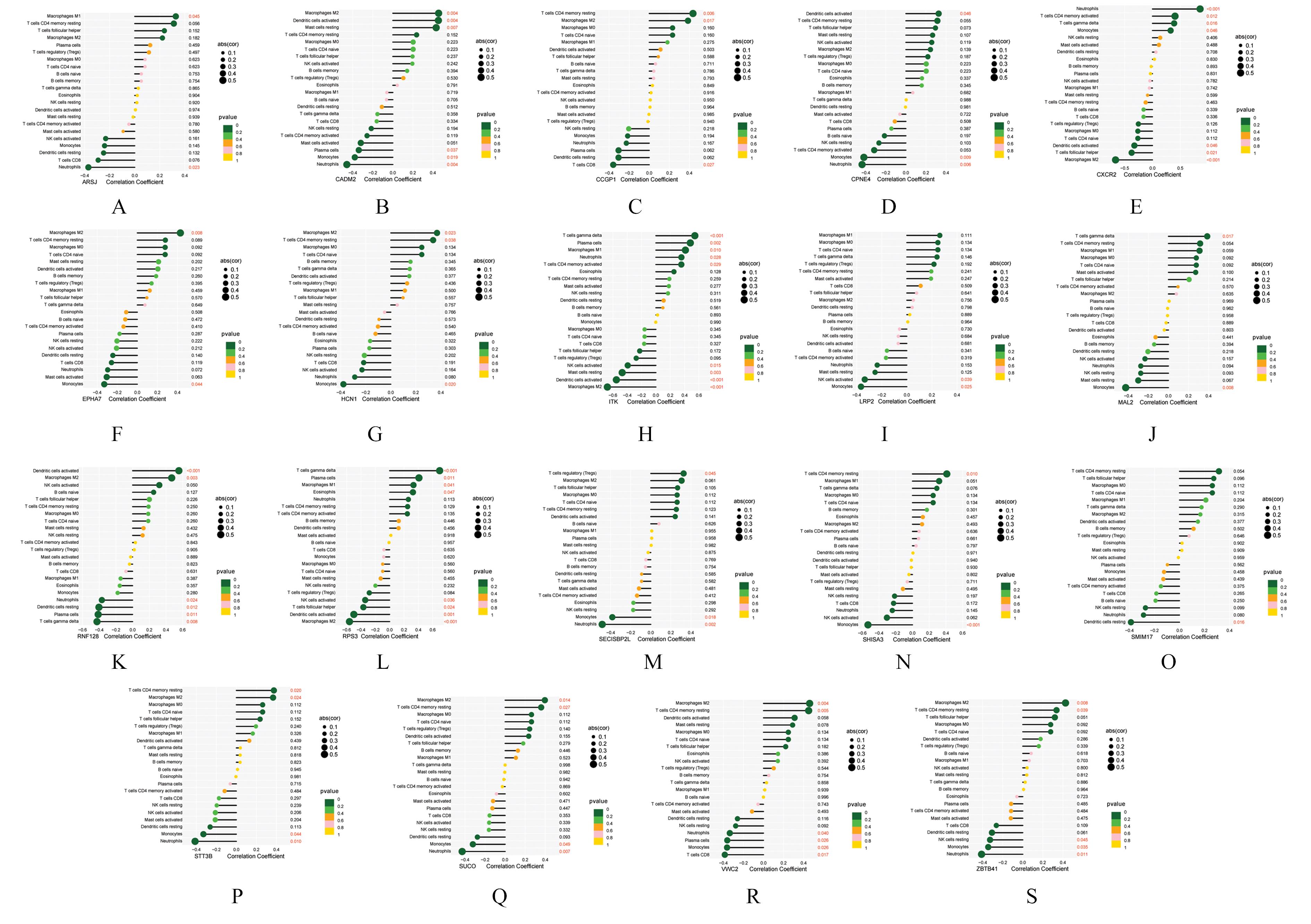
Tab.1
Interaction among circRNA, miRNA, mRNA and their target genes in ceRNA network"
| circRNA | miRNA | mRNA | circFC | mFC | circRNA | miRNA | mRNA | circFC | mFC |
|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0017037 | hsa-miR-142-5p | MAL2 | -2.50 | -1.48 | hsa_circ_0024957 | hsa-miR-142-5p | SHISA3 | -2.66 | -1.32 |
| hsa_circ_0017037 | hsa-miR-142-5p | STT3B | -2.50 | -1.09 | hsa_circ_0024957 | hsa-miR-142-5p | ZBTB41 | -2.66 | -1.09 |
| hsa_circ_0017037 | hsa-miR-142-5p | SHISA3 | -2.50 | -1.32 | hsa_circ_0024957 | hsa-miR-142-5p | CPNE4 | -2.66 | -1.63 |
| hsa_circ_0017037 | hsa-miR-142-5p | ZBTB41 | -2.50 | -1.09 | hsa_circ_0024957 | hsa-miR-142-5p | SMIM17 | -2.66 | -1.11 |
| hsa_circ_0017037 | hsa-miR-142-5p | CPNE4 | -2.50 | -1.63 | hsa_circ_0024957 | hsa-miR-142-5p | CADM2 | -2.66 | -1.07 |
| hsa_circ_0017037 | hsa-miR-142-5p | SMIM17 | -2.50 | -1.11 | hsa_circ_0024957 | hsa-miR-142-5p | RNF128 | -2.66 | -1.92 |
| hsa_circ_0017037 | hsa-miR-142-5p | CADM2 | -2.50 | -1.07 | hsa_circ_0024957 | hsa-miR-142-5p | LRP2 | -2.66 | -1.43 |
| hsa_circ_0017037 | hsa-miR-142-5p | RNF128 | -2.50 | -1.92 | hsa_circ_0024957 | hsa-miR-142-5p | ARSJ | -2.66 | -1.23 |
| hsa_circ_0017037 | hsa-miR-142-5p | LRP2 | -2.50 | -1.43 | hsa_circ_0024957 | hsa-miR-142-5p | CCPG1 | -2.66 | -1.17 |
| hsa_circ_0017037 | hsa-miR-142-5p | ARSJ | -2.50 | -1.23 | hsa_circ_0024957 | hsa-miR-199a-5p | EPHA7 | -2.66 | -1.11 |
| hsa_circ_0017037 | hsa-miR-142-5p | CCPG1 | -2.50 | -1.17 | hsa_circ_0024957 | hsa-miR-199a-5p | SUCO | -2.66 | -1.16 |
| hsa_circ_0006562 | hsa-miR-142-5p | MAL2 | -2.63 | -1.48 | hsa_circ_0024957 | hsa-miR-199a-3p | SECISBP2L | -2.66 | -1.01 |
| hsa_circ_0006562 | hsa-miR-142-5p | STT3B | -2.63 | -1.09 | hsa_circ_0024957 | hsa-miR-199a-3p | LRP2 | -2.66 | -1.43 |
| hsa_circ_0006562 | hsa-miR-142-5p | SHISA3 | -2.63 | -1.32 | hsa_circ_0043598 | hsa-miR-199a-5p | EPHA7 | -3.53 | -1.11 |
| hsa_circ_0006562 | hsa-miR-142-5p | ZBTB41 | -2.63 | -1.09 | hsa_circ_0043598 | hsa-miR-199a-5p | SUCO | -3.53 | -1.16 |
| hsa_circ_0006562 | hsa-miR-142-5p | CPNE4 | -2.63 | -1.63 | hsa_circ_0062993 | hsa-miR-142-3p | SUCO | -2.03 | -1.16 |
| hsa_circ_0006562 | hsa-miR-142-5p | SMIM17 | -2.63 | -1.11 | hsa_circ_0062993 | hsa-miR-142-3p | ZBTB41 | -2.03 | -1.09 |
| hsa_circ_0006562 | hsa-miR-142-5p | CADM2 | -2.63 | -1.07 | hsa_circ_0062993 | hsa-miR-142-3p | VWC2 | -2.03 | -1.21 |
| hsa_circ_0006562 | hsa-miR-142-5p | RNF128 | -2.63 | -1.92 | hsa_circ_0062993 | hsa-miR-142-3p | HCN1 | -2.03 | -1.05 |
| hsa_circ_0006562 | hsa-miR-142-5p | LRP2 | -2.63 | -1.43 | hsa_circ_0002021 | hsa-miR-193a-5p | ITK | 2.14 | 1.08 |
| hsa_circ_0006562 | hsa-miR-142-5p | ARSJ | -2.63 | -1.23 | hsa_circ_0002021 | hsa-miR-193a-5p | CXCR2 | 2.14 | 2.46 |
| hsa_circ_0006562 | hsa-miR-142-5p | CCPG1 | -2.63 | -1.17 | hsa_circ_0002021 | hsa-miR-193a-5p | RPS3 | 2.14 | 1.24 |
| hsa_circ_0024957 | hsa-miR-142-5p | MAL2 | -2.66 | -1.48 | hsa_circ_0055451 | hsa-miR-199a-3p | SECISBP2L | -4.26 | -1.01 |
| hsa_circ_0024957 | hsa-miR-142-5p | STT3B | -2.66 | -1.09 | hsa_circ_0055451 | hsa-miR-199a-3p | LRP2 | -4.26 | -1.43 |
| 1 | XIAO Y, WANG X Y, YANG J, et al. Huatan Dingji Decoction intervening in atrial fibrillation: protocol for a randomized double-blind single-simulated placebo-controlled clinical trial[J]. Trials, 2021, 22(1): 693. |
| 2 | LIU Y Z, SHI Q M, MA Y X, et al. The role of immune cells in atrial fibrillation[J]. J Mol Cell Cardiol, 2018, 123: 198-208. |
| 3 | CAO F, LI Z, DING W M,et al.Angiotensin Ⅱ-treated cardiac myocytes regulate M1 macrophage polarization via transferring exosomal PVT1[J]. J Immunol Res, 2021, 2021: 1994328. |
| 4 | MIYOSAWA K, IWATA H, MINAMI-TAKANO A, et al. Enhanced monocyte migratory activity in the pathogenesis of structural remodeling in atrial fibrillation[J]. PLoS One, 2020, 15(10): e0240540. |
| 5 | SZABO L, SALZMAN J. Detecting circular RNAs: bioinformatic and experimental challenges[J]. Nat Rev Genet, 2016, 17(11): 679-692. |
| 6 | BARRETT S P, SALZMAN J. Circular RNAs: analysis, expression and potential functions[J]. Development, 2016, 143(11): 1838-1847. |
| 7 | ZHANG X H, LU J Y, ZHANG Q H, et al. CircRNA RSF1 regulated ox-LDL induced vascular endothelial cells proliferation, apoptosis and inflammation through modulating miR-135b-5p/HDAC1 axis in atherosclerosis[J]. Biol Res, 2021, 54(1): 11. |
| 8 | TIAN T T, LI F, CHEN R H, et al. Therapeutic potential of exosomes derived from circRNA_0002113 lacking mesenchymal stem cells in myocardial infarction[J]. Front Cell Dev Biol, 2022, 9: 779524. |
| 9 | WANG K, LONG B, LIU F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223[J]. Eur Heart J, 2016, 37(33): 2602-2611. |
| 10 | SU Q, LV X W. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis[J]. Genomics, 2020, 112(2): 1680-1685. |
| 11 | WU N, LI C Y, XU B, et al. Circular RNA mmu_circ_0005019 inhibits fibrosis of cardiac fibroblasts and reverses electrical remodeling of cardiomyocytes[J]. BMC Cardiovasc Disord, 2021, 21(1): 308. |
| 12 | RITCHIE M E, PHIPSON B, WU D, et al. Limma Powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Res, 2015, 43(7): e47. |
| 13 | GHOSAL S, DAS S, SEN R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits[J]. Front Genet, 2013, 4: 283. |
| 14 | LI J H, LIU S, ZHOU H, et al. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data[J]. Nucl Acids Res, 2014, 42(D1): D92-D97. |
| 15 | LIU M, WANG Q, SHEN J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature[J]. RNA Biol, 2019, 16(7): 899-905. |
| 16 | AGARWAL V, BELL G W, NAM J W, et al. Predicting effective microRNA target sites in mammalian mRNAs[J]. eLife, 2015, 4: e05005. |
| 17 | WONG N, WANG X W.miRDB: an online resource for microRNA target prediction and functional annotations[J].Nucleic Acids Res,2015,43(Database issue): D146-D152. |
| 18 | HUANG D W, SHERMAN B T, LEMPICKI R A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009, 4(1): 44-57. |
| 19 | NEWMAN A M, LIU C L, GREEN M R, et al. Robust enumeration of cell subsets from tissue expression profiles[J].Nat Methods,2015,12(5): 453-457. |
| 20 | KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691. |
| 21 | ZHANG Y L, TENG F, HAN X, et al. Selective blocking of CXCR2 prevents and reverses atrial fibrillation in spontaneously hypertensive rats[J]. J Cell Mol Med, 2020, 24(19): 11272-11282. |
| 22 | GROSSI I, SALVI A, ABENI E, et al. Biological function of microRNA193a-3p in health and disease[J]. Int J Genomics, 2017, 2017: 5913195. |
| 23 | HELLER G, WEINZIERL M, NOLL C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers[J]. Clin Cancer Res, 2012, 18(6): 1619-1629. |
| 24 | ZHAN L Y, LEI S Q, LI W L, et al. Suppression of microRNA-142-5p attenuates hypoxia-induced apoptosis through targeting SIRT7[J]. Biomed Pharmacother, 2017, 94: 394-401. |
| 25 | WANG A, KWEE L C, GRASS E, et al. Whole blood sequencing reveals circulating microRNA associations with high-risk traits in non-ST-segment elevation acute coronary syndrome[J]. Atherosclerosis, 2017, 261: 19-25. |
| 26 | ZARJOU A, YANG S Z, ABRAHAM E, et al. Identification of a microRNA signature in renal fibrosis: role of miR-21[J]. Am J Physiol Renal Physiol, 2011, 301(4): F793-F801. |
| 27 | WANG H, WANG Z X, TANG Q B. Reduced expression of microRNA-199a-3p is associated with vascular endothelial cell injury induced by type 2 diabetes mellitus[J]. Exp Ther Med, 2018, 16(4): 3639-3645. |
| 28 | DENHAM N C, PEARMAN C M, CALDWELL J L, et al. Calcium in the pathophysiology of atrial fibrillation and heart failure[J]. Front Physiol, 2018, 9: 1380. |
| 29 | URBANTAT R M, VAJKOCZY P, BRANDENBURG S. Advances in chemokine signaling pathways as therapeutic targets in glioblastoma[J]. Cancers, 2021, 13(12): 2983. |
| 30 | LIU P, SUN H K, ZHOU X, et al. CXCL12/CXCR4 axis as a key mediator in atrial fibrillation via bioinformatics analysis and functional identification[J]. Cell Death Dis, 2021, 12(9): 813. |
| 31 | KAWASAKI M, MEULENDIJKS E R, VAN DEN BERG N W E, et al. Neutrophil degranulation interconnects over-represented biological processes in atrial fibrillation[J]. Sci Rep, 2021, 11(1): 2972. |
| 32 | ERTAS G, SÖNMEZ O, TURFAN M,et al. Neutrophil/lymphocyte ratio is associated with thromboembolic stroke in patients with non-valvular atrial fibrillation[J]. J Neurol Sci, 2013, 324(1/2): 49-52. |
| 33 | LI S B, YANG F, JING L, et al. Myeloperoxidase and risk of recurrence of atrial fibrillation after catheter ablation[J]. J Investig Med, 2013, 61(4): 722-727. |
| [1] | Ying ZHAO,Danyu ZHAO,Chao LIU. Bioinformatics analysis on prognostic evaluation value of TXNDC11 gene in pan-cancer and its immunity regulation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 142-153. |
| [2] | Lan ZHENG,Songzhe PIAO,Ran XU,Xinyue WANG,Yixuan WANG,Zhenhua LIN,Yang YANG. Bioinformatics analysis based on effect of expression levels of m6A regulators on immune infiltration and prognosis of uterine corpus endometrial carcinoma [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 438-452. |
| [3] | Ye YUAN,Jinbao ZHUANG,Xu SHI,Changyuan LI. Influence of PGE2 on PD-1 expression in infiltrating T lymphocytes in non-small cell lung cancer tissue and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2021, 47(2): 249-256. |
| [4] | LI Xiaobing, WANG Xu, LYU Ying, HUANG Jiancheng, LI Hongying, WANG Jun, ZHANG Huijun, SU Zhenyu. Effects of salvianolic acid on levels of VCAM-1 and ICAM-1 in myocardium tissue and ERK1/2-NF-κB signaling pathway of atrial fibrillation rats [J]. Journal of Jilin University(Medicine Edition), 2020, 46(05): 1004-1010. |
| [5] | WANG Zhi, HONG Li, LI Suting, ZENG Wanling. Analysis on endometrial cancer-related genes and candidate pathways based on GEO database bioinformatics methods [J]. Journal of Jilin University(Medicine Edition), 2020, 46(04): 804-809. |
| [6] | SHI Hui, ZHAO Lulu, DU Yunhui, HUA Baotong, GUO Tao. Changes of RAAS in atrial fibrillation model dogs after spinal cord stimulation and its inhibitory effect on atrial fibrillation [J]. Journal of Jilin University(Medicine Edition), 2019, 45(03): 511-517. |
| [7] | WANG Tianyue, ZHOU Qianlan, SHANG Yunxiao. Effect of epigallocatechin-3-gallate on airway inflammation and Treg/Th17 immune balance of mice with obese asthma [J]. Journal of Jilin University(Medicine Edition), 2019, 45(03): 491-497. |
| [8] | PAN Junqiang, ZHANG Dianxin, SUN Chaofeng, YANG Jin. Expression of fibrillin-1 in atrium tissue and analysis on its relationship with atrial fibrosis in patients with valvular atrial fibrillation [J]. Journal of Jilin University Medicine Edition, 2017, 43(02): 339-343. |
| [9] | LA Zong,WANG Jian-xia,CUI Ni,LI Ming-yue,XING Shu-gang,REN Bo,ZHANG Li-hong,LI Wei,LI Yu-lin. Correlation of CD4 and CD8 positive lymphocytes with tumor angiogenesis in microenvironment of breast invasive ductal carcinoma [J]. Journal of Jilin University Medicine Edition, 2014, 40(05): 1064-1073. |
| [10] | LI Xi, XIE Ying, ZHANG Wei-jun,WEN Shao-jun, ZHAO Rui-xiang, PENG Xin-jie, ZHANG Wen, ZHANG Yan, CHENG Xiu. Changes of coagulation and fibrinolysis in middle-old aged patients with nonvalvular atrial fibrillation [J]. J4, 2006, 32(3): 461-464. |
| [11] | SUN Yan-bo, SHI Hong-yan, LI Jing-hua. Construction of prokaryotic expression vector [J]. J4, 2006, 32(3): 373-375. |
| [12] | MA Rong,WU Ying-quan,JIA Jian-ping. Changes of peripheral blood cortisol lymphocytes and T lymphocytes subsets in patients with acute cerebrovascular diseases in state of stress [J]. J4, 2005, 31(4): 580-583. |
| [13] | CAO Jie, SUN Lian-kun, CHENG Xiao-geng, MAO Jian-hua,LIU Xi-chun, WANG Cheng, LI Yang, ZHAO Xue-jian. Effects of IFN-PDS on T-lymphocyte proliferationin influenza virus-infected mice [J]. J4, 2004, 30(3): 357-359. |
|