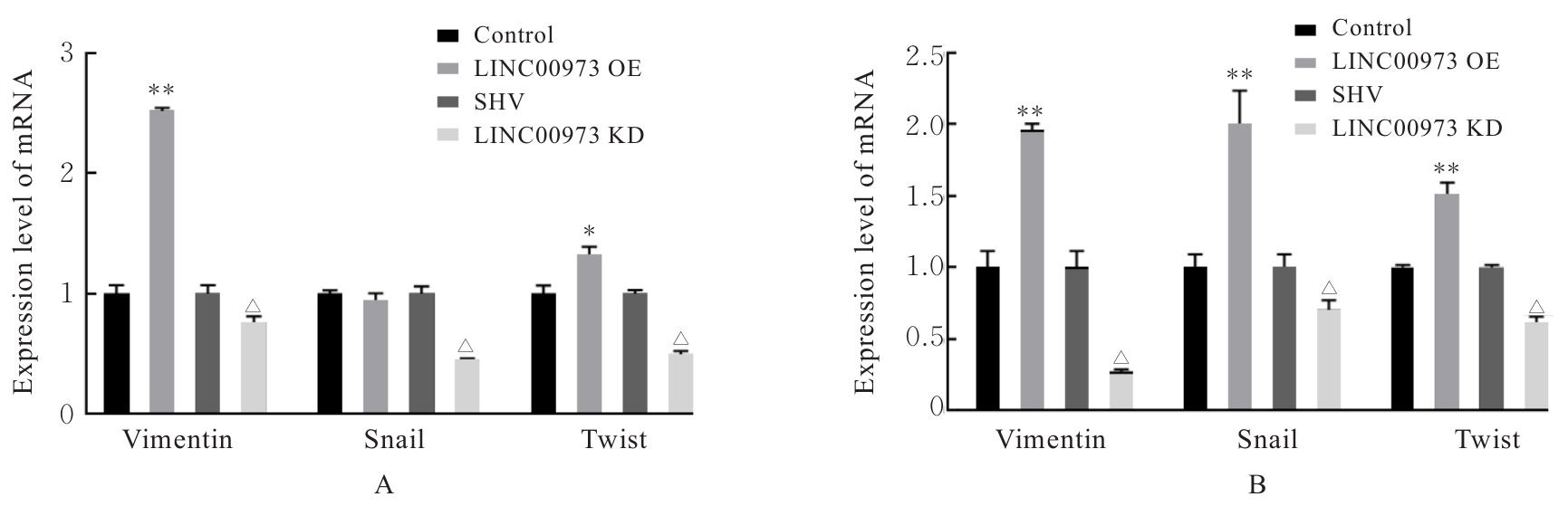
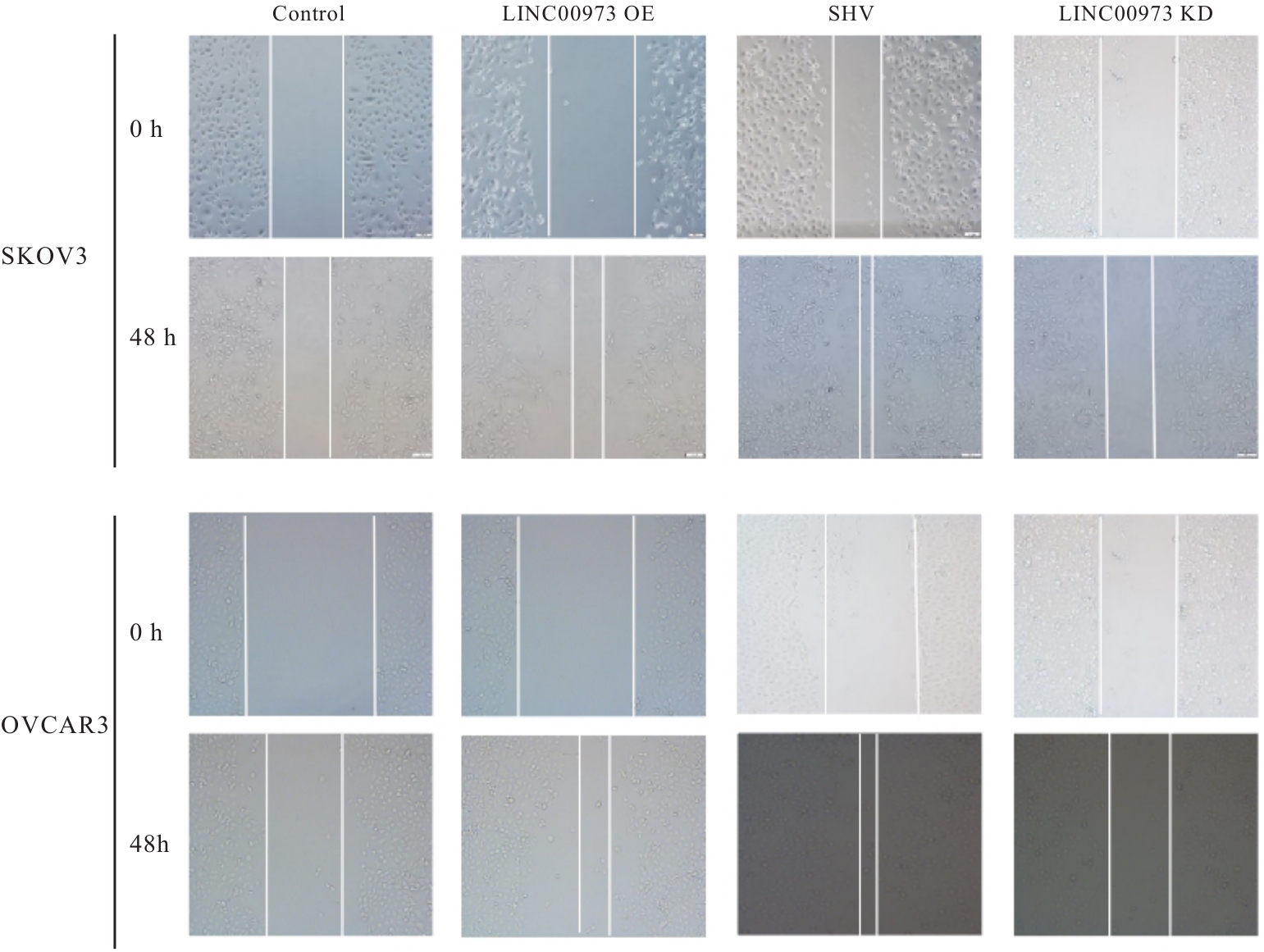
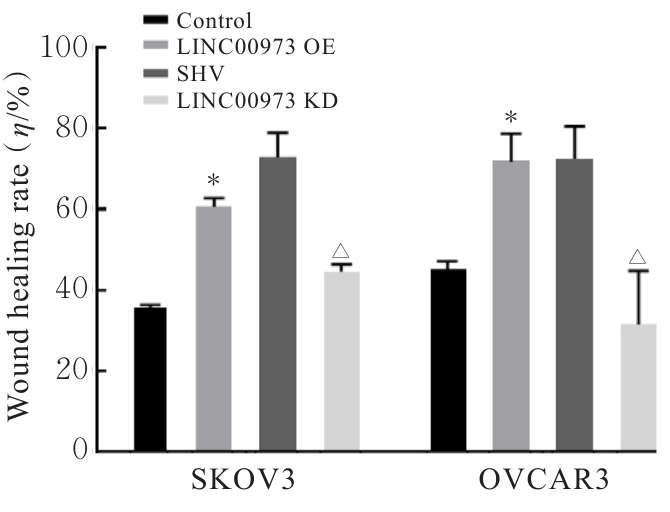
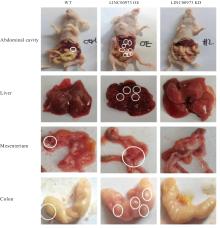
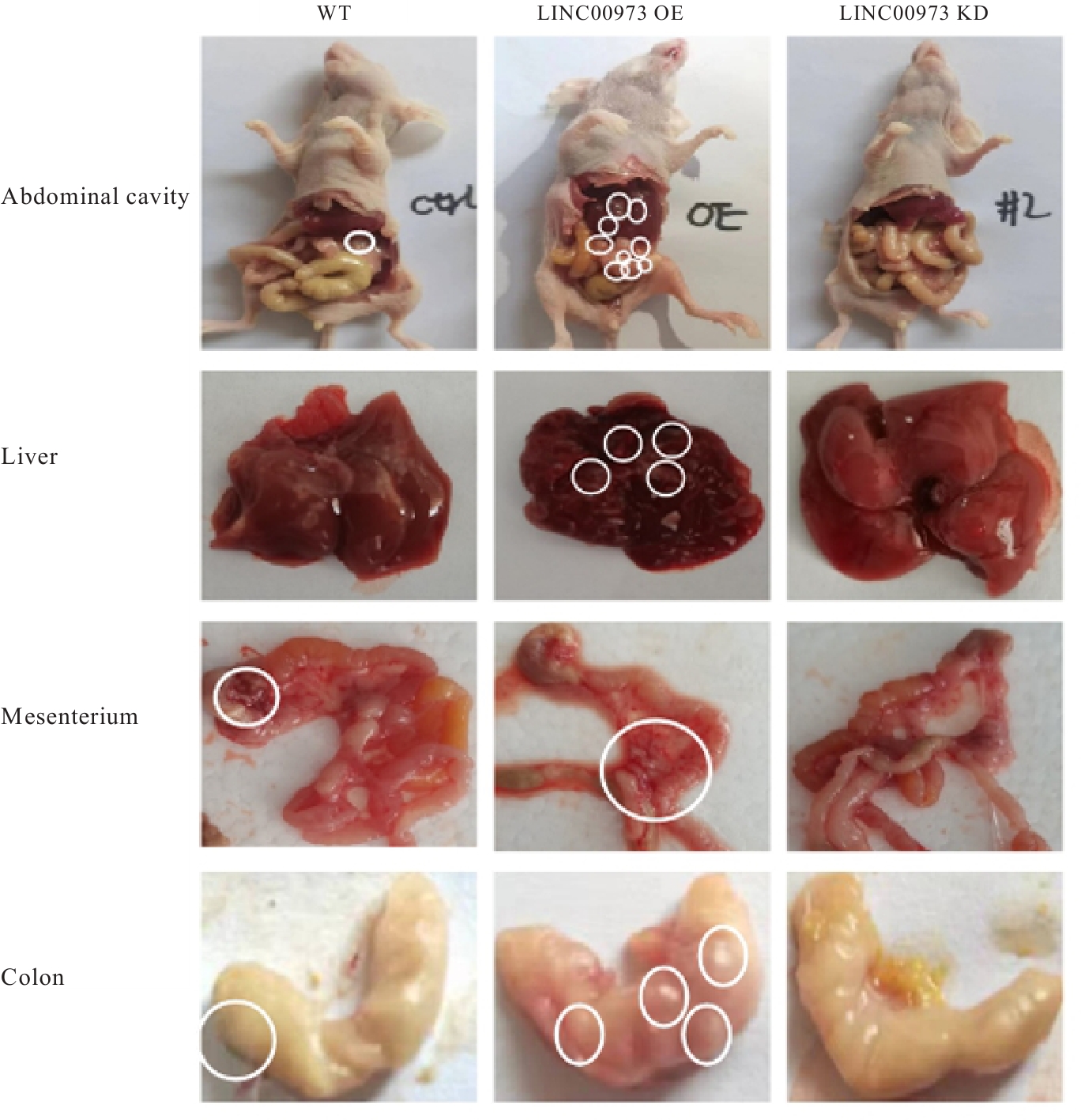
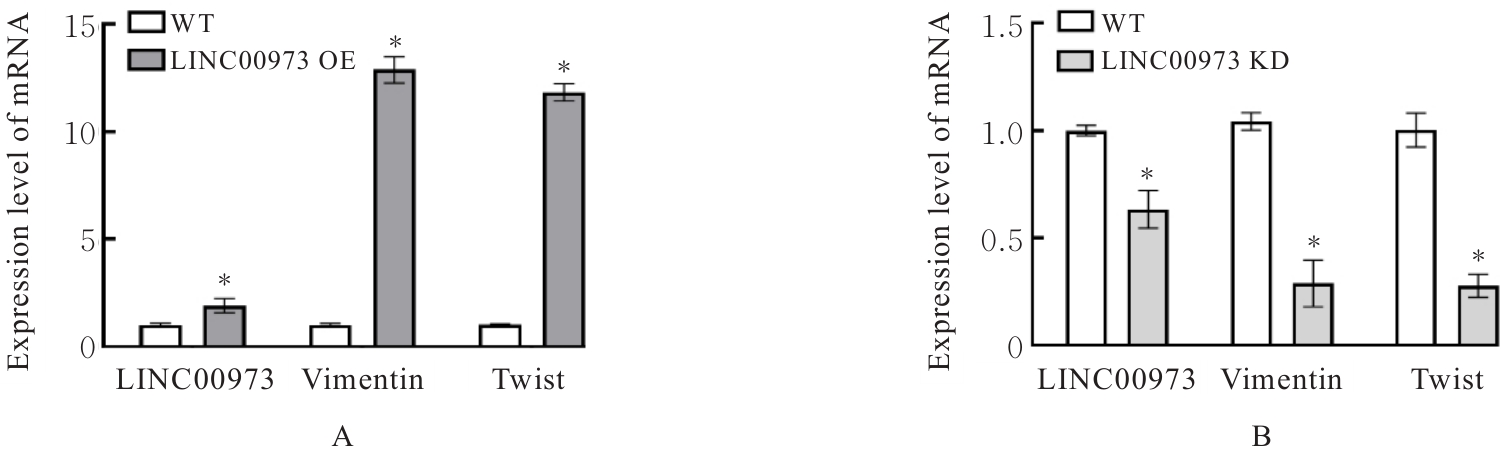
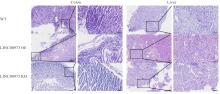
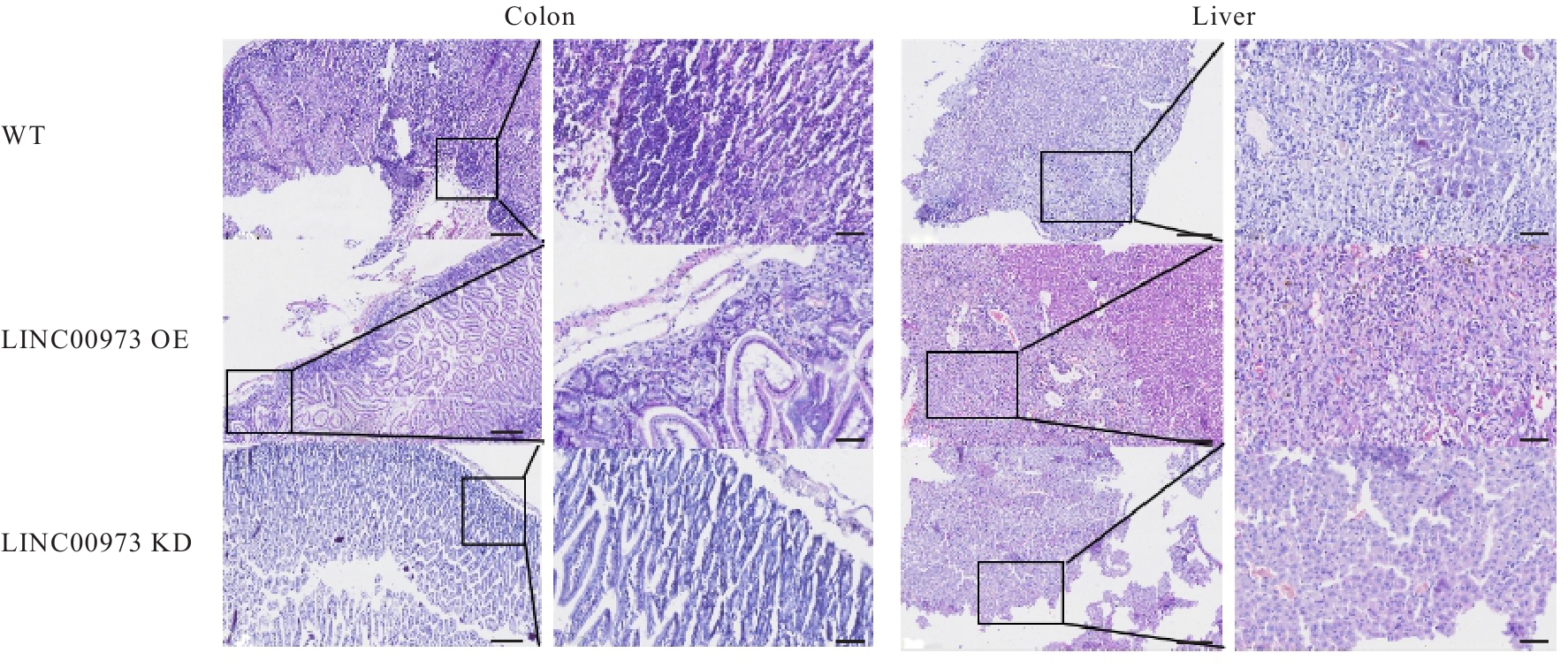
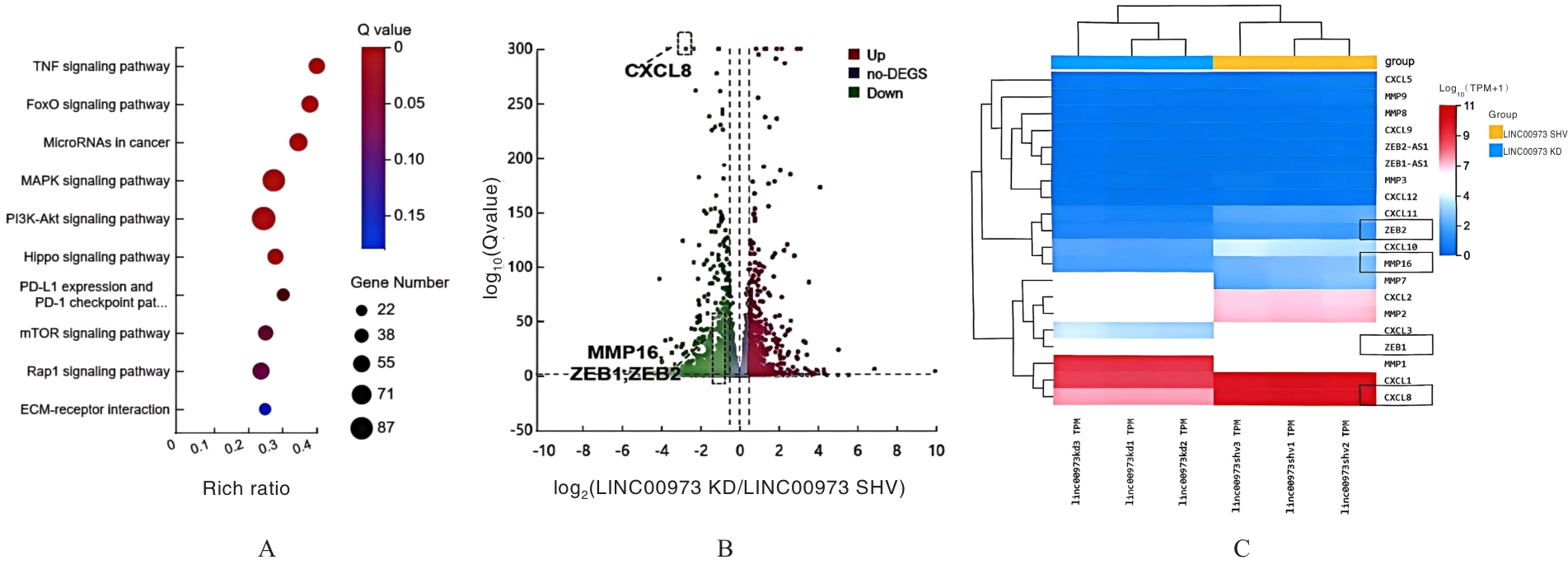
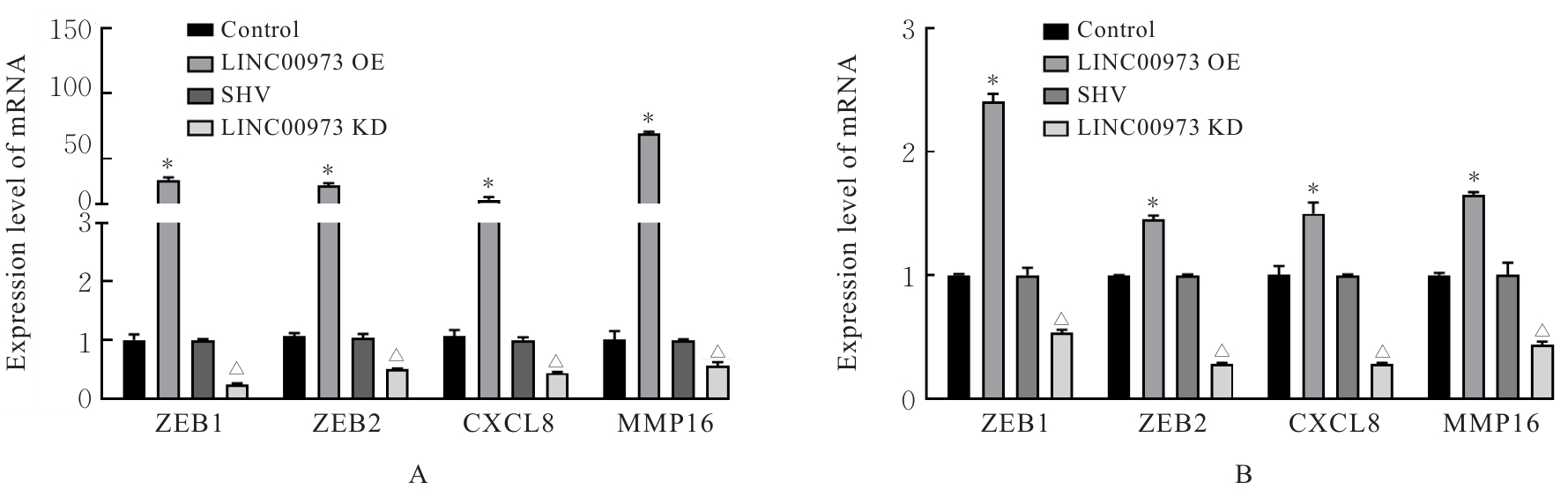
Journal of Jilin University(Medicine Edition) ›› 2025, Vol. 51 ›› Issue (4): 866-878.doi: 10.13481/j.1671-587X.20250402
• Research in basic medicine • Previous Articles Next Articles
Long non-coding RNA LINC00973 promotes migration, invasion and distal metastasis of epithelial ovarian cancer and its molecular mechanism
Yunxiu XIA1,2,Shuo ZHANG1,2,Huanhai ZHANG3,Fei WANG2,Hongliang DONG2( ),Jing DU1,2(
),Jing DU1,2( )
)
- 1.Department of Gynecology,Affiliated Hospital,Binzhou Medical University,Binzhou 256603,China
2.Medical Research Center,Affiliated Hospital,Binzhou Medical University,Binzhou 256603,China
3.Department of Thyroid and Breast Surgery,Yantai Affiliated Hospital,Binzhou Medical University,Yantai 264100,China
-
Received:2024-09-08Accepted:2024-10-24Online:2025-07-28Published:2025-08-25 -
Contact:Hongliang DONG,Jing DU E-mail:hongliang.234@163.com;djedith@126.com
CLC Number:
- R737.31
Cite this article
Yunxiu XIA,Shuo ZHANG,Huanhai ZHANG,Fei WANG,Hongliang DONG,Jing DU. Long non-coding RNA LINC00973 promotes migration, invasion and distal metastasis of epithelial ovarian cancer and its molecular mechanism[J].Journal of Jilin University(Medicine Edition), 2025, 51(4): 866-878.
share this article
Tab. 1
Sequences of primers"
| Gene | Primer(5'-3') |
|---|---|
| LINC00973 | F: CAAGGACAATTATTCCCTCAC R: GACAAAGCCAAGGATTCAGT |
| Vimentin | F: CGCCAACTACATCGACAAGGTGC R: CTGGTCCACCTGCCGGCGCAG |
| Snail | F: TCCACGAGGTGTGACTAACT R: CCGACAAGTGACAGCCATTA |
| Twist | F: CTAGAGACTCTGGAGCTGGATA R: TCTGGGAATCACTGTCCACG |
| ZEB1 | F: AAGTGGCGGTAGATGGTA R: TGTTGTATGGGTGAAGCA |
| ZEB2 | F: TCTGCGACATAAATACGA R: GAGTGAAGCCTTGAGTGC |
| MMP16 | F: TGTGATGGACCAACAGAC R: ATACAAGAGGCATAAGG |
| CXCL8 | F: CTAGTTTGATACTCCCAGTC R: AAGTTTCAACCAGCAAGA |
| GAPDH | F: CTCCTCCTGTTCGACAGTCAGC R: CCCAATACGACCAAATCCGTT |
| [1] | WEBB P M, JORDAN S J. Global epidemiology of epithelial ovarian cancer[J]. Nat Rev Clin Oncol, 2024, 21(5): 389-400. |
| [2] | TORRE L A, TRABERT B, DESANTIS C E, et al. Ovarian cancer statistics, 2018[J]. CA A Cancer J Clin, 2018, 68(4): 284-296. |
| [3] | BRIDGES M C, DAULAGALA A C, KOURTIDIS A. LNCcation: lncRNA localization and function[J]. J Cell Biol, 2021, 220(2): e202009045. |
| [4] | ZHANG Q, WU E Z, TANG Y H, et al. Deeply mining a universe of peptides encoded by long noncoding RNAs[J]. Mol Cell Proteomics, 2021, 20: 100109. |
| [5] | HERMAN A B, TSITSIPATIS D, GOROSPE M. Integrated lncRNA function upon genomic and epigenomic regulation[J]. Mol Cell, 2022, 82(12): 2252-2266. |
| [6] | HE S L, CHEN Y L, CHEN Q H, et al. LncRNA KCNQ1OT1 promotes the metastasis of ovarian cancer by increasing the methylation of EIF2B5 promoter[J]. Mol Med, 2022, 28(1): 112. |
| [7] | YU X, ZHAO P F, LUO Q Y, et al. RUNX1-IT1 acts as a scaffold of STAT1 and NuRD complex to promote ROS-mediated NF-κB activation and ovarian cancer progression[J]. Oncogene, 2024, 43(6): 420-433. |
| [8] | LIN H, XU X, CHEN K L, et al. LncRNA CASC15, miR-23b cluster and SMAD3 form a novel positive feedback loop to promote epithelial-mesenchymal transition and metastasis in ovarian cancer[J]. Int J Biol Sci, 2022, 18(5): 1989-2002. |
| [9] | KARPOV D S, SPIRIN P V, ZHELTUKHIN A O, et al. LINC00973 induces proliferation arrest of drug-treated cancer cells by preventing p21 degradation[J]. Int J Mol Sci, 2020, 21(21): 8322. |
| [10] | ZINOVIEVA O L, GRINEVA E N, PROKOFJEVA M M, et al. Expression of long non-coding RNA LINC00973 is consistently increased upon treatment of colon cancer cells with different chemotherapeutic drugs[J]. Biochimie, 2018, 151: 67-72. |
| [11] | LIU Y B, LI X Z, ZHANG C M, et al. LINC00973 is involved in cancer immune suppression through positive regulation of Siglec-15 in clear-cell renal cell carcinoma[J]. Cancer Sci, 2020, 111(10): 3693-3704. |
| [12] | GUO Q, LI D, LUO X Y, et al. The regulatory network and potential role of LINC00973-miRNA-mRNA ceRNA in the progression of non-small-cell lung cancer[J]. Front Immunol, 2021, 12: 684807. |
| [13] | TADIĆ V, ZHANG W, BROZOVIC A. The high-grade serous ovarian cancer metastasis and chemoresistance in 3D models[J]. Biochim Biophys Acta Rev Cancer, 2024, 1879(1): 189052. |
| [14] | CAI Y S, LYU T, LI H, et al. LncRNA CEBPA-DT promotes liver cancer metastasis through DDR2/β-catenin activation via interacting with hnRNPC[J]. J Exp Clin Cancer Res, 2022, 41(1): 335. |
| [15] | XIU B Q, CHI Y Y, LIU L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription[J]. Mol Cancer, 2019, 18(1): 187. |
| [16] | WANG Y, LIU F, CHEN L, et al. Neutrophil extracellular traps (NETs) promote non-small cell lung cancer metastasis by suppressing lncRNA MIR503HG to activate the NF-κB/NLRP3 inflammasome pathway[J]. Front Immunol, 2022, 13: 867516. |
| [17] | HU H F, HAN L, FU J Y, et al. LINC00982-encoded protein PRDM16-DT regulates CHEK2 splicing to suppress colorectal cancer metastasis and chemoresistance[J]. Theranostics, 2024, 14(8): 3317-3338. |
| [18] | TONG L L, WANG Y Y, AO Y, et al. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis[J]. Biomed Pharmacother, 2019, 115: 108891. |
| [19] | LIU J, ZHU Y, WANG H, et al. LINC00629, a HOXB4-downregulated long noncoding RNA, inhibits glycolysis and ovarian cancer progression by destabilizing c-Myc[J]. Cancer Sci, 2024, 115(3): 804-819. |
| [20] | LI Y, LOU S H, ZHANG J, et al. m6A methylation-mediated regulation of LncRNA MEG3 suppresses ovarian cancer progression through miR-885-5p and the VASH1 pathway[J]. J Transl Med, 2024, 22(1): 113. |
| [21] | FARDI M, ALIVAND M, BARADARAN B, et al. The crucial role of ZEB2: from development to epithelial-to-mesenchymal transition and cancer complexity[J]. J Cell Physiol, 2019, 234(9): 14783-14799. |
| [22] | SáNCHEZ-TILLó E, PEDROSA L, VILA I, et al. The EMT factor ZEB1 paradoxically inhibits EMT in BRAF-mutant carcinomas[J]. JCI Insight, 2023, 8(20):e164629. |
| [23] | POULIQUEN D L, BOISSARD A, HENRY C, et al. Curcuminoids as modulators of EMT in invasive cancers: a review of molecular targets with the contribution of malignant mesothelioma studies[J]. Front Pharmacol, 2022, 13: 934534. |
| [24] | SUN Z Q, SHAO B, LIU Z Q, et al. LINC01296/miR-141-3p/ZEB1-ZEB2 axis promotes tumor metastasis via enhancing epithelial-mesenchymal transition process[J]. J Cancer, 2021, 12(9): 2723-2734. |
| [25] | LI Y Y, FEI H, LIN Q W, et al. ZEB2 facilitates peritoneal metastasis by regulating the invasiveness and tumorigenesis of cancer stem-like cells in high-grade serous ovarian cancers[J]. Oncogene, 2021, 40(32): 5131-5141. |
| [26] | TATTI O, GUCCIARDO E, PEKKONEN P, et al. MMP16 mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma[J]. Cancer Res, 2015, 75(10): 2083-2094. |
| [27] | YAN P F, WANG J, LIU H Y, et al. M1 macrophage-derived exosomes containing miR-150 inhibit glioma progression by targeting MMP16[J]. Cell Signal, 2023, 108: 110731. |
| [28] | YANG F Q, ZHANG J Q, JIN J J, et al. HOXA11-AS promotes the growth and invasion of renal cancer by sponging miR-146b-5p to upregulate MMP16 expression[J]. J Cell Physiol, 2018, 233(12): 9611-9619. |
| [29] | LI Y Y, WANG Y, YU L, et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16[J]. Cancer Lett, 2013, 339(2): 260-269. |
| [30] | LIU Q, LI A P, TIAN Y J, et al. The CXCL8-CXCR1/2 pathways in cancer[J]. Cytokine Growth Factor Rev, 2016, 31: 61-71. |
| [31] | PIAO H Y, FU L F, WANG Y X, et al. A positive feedback loop between gastric cancer cells and tumor-associated macrophage induces malignancy progression[J]. J Exp Clin Cancer Res, 2022, 41(1): 174. |
| [32] | FU X R, WANG Q M, DU H, et al. CXCL8 and the peritoneal metastasis of ovarian and gastric cancer[J]. Front Immunol, 2023, 14: 1159061. |
| [33] | ZHANG N, LIU Y Y, WANG Y Y, et al. Decitabine reverses TGF-β1-induced epithelial-mesenchymal transition in non-small-cell lung cancer by regulating miR-200/ZEB axis[J]. Drug Des Devel Ther, 2017, 11: 969-983. |
| [34] | CHOI P W, NG S W. The functions of microRNA-200 family in ovarian cancer: beyond epithelial-mesenchymal transition[J]. Int J Mol Sci, 2017, 18(6): 1207. |
| [35] | ZHAO M N, ZHANG L F, SUN Z, et al. A novel microRNA-182/Interleukin-8 regulatory axis controls osteolytic bone metastasis of lung cancer[J]. Cell Death Dis, 2023, 14(5): 298. |
| [1] | Shuo ZHANG,Yunxiu XIA,Weiwei CHEN,Hongliang DONG,Bingjie CUI,Cuilan LIU,Zhiqiang LIU,Fei WANG,Jing DU. Effect of over-expression of NR2F2 on biological behaviors of human ovarian cancer SKOV3 cells [J]. Journal of Jilin University(Medicine Edition), 2025, 51(1): 58-67. |
| [2] | Jia ZHOU,Zhidong QIU,Zhe LIN,Guangfu LYU,Jiaming XU,He LIN,Kexin WANG,Yuchen WANG,Xiaowei HUANG. Effect of chelerythrine on migration, invasion, and epithelial-mesenchymal transition of human ovarian cancer SKOV3 cells [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 25-32. |
| [3] | Shaoting HUANG,You LI,Zhaochun WU,Jiawen HE,Keqi LIAO,Shengnan LI. Construction of SOX17 over-expression lentiviral vector and stably transfected cell line [J]. Journal of Jilin University(Medicine Edition), 2023, 49(6): 1424-1430. |
| [4] | Jingjing LI, REZIWANGULI·Aisikaier,Wanyi XING,Yinggang ZOU. Primary dedifferentiated liposarcoma of unilateral ovary: A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 1040-1045. |
| [5] | Mengmeng CHEN,Junyu CHEN,Yan GAO,Zhengxuan FANG,Shuhua ZHAO,Jingying ZHENG,Shuxiang LIU. Expression level of gasdermin E in epithelial ovarian cancer tissue and its clinical significance [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 440-451. |
| [6] | Mengshi MA,Peng CHEN,Zhongsen MA. Pulmonary benign metastasizing leiomyoma:A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 203-208. |
| [7] | Yanan MA,Bing TAN,Lei XU,Jiandong ZHANG. Detection of drug resistance genes of Acinetobacter baumannii in sputum samples of ICU patients by real-time fluorescence quantitative PCR method and its evaluation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1623-1628. |
| [8] | Qinghui WANG,Bo LI,Chuancui HU,Mingchao NIE,Xiaomei ZHENG. Effects of miR-107 on regulation of immune escape and taxol resistance of ovarian cancer cells [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 734-743. |
| [9] | Shuqing FENG,Yanhong YAO,Yue SHI,Zhibin LIU,Feng GAO. Application of real-time fluorescence quantitative PCR for detection of BCR/ABL gene expression in peripheral blood in hematopoietic stem cell transplantation therapy in patients with Ph + acute lymphoblastic leukemia [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 180-187. |
| [10] | Jingjing GE,Hongxia XU,Lixia XIE,Kaili DU,Ming SANG,Xiaodong SUN. Establishment of human serous ovarian cancer cell culture system in vitro and its application in chemotherapeutic drug sensitivity detection [J]. Journal of Jilin University(Medicine Edition), 2021, 47(4): 999-1007. |
| [11] | GUO Xiaofei, LIU Lihong, AI Hao. Expression of valosin-containing protein in epithelial ovarian cancer tissue and its clinical significance [J]. Journal of Jilin University(Medicine Edition), 2020, 46(03): 600-606. |
| [12] | PEI Yanling, LYU Hanghang, WANG Bowei, ZHANG Weiyang. Misdiagnosis of ovarian fibrothecoma as ovarian malignant tumor: A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2019, 45(04): 940-943. |
| [13] | WANG Yipin, HE Dongyun, HUANG Qiongwei, SHENG Minjia. Application value of risk ovarian malignancy algorithm in early diagnosis of epithelial ovarian cancer [J]. Journal of Jilin University(Medicine Edition), 2019, 45(04): 905-910. |
| [14] | XING Shugang, ZHANG Lihong, JIN Minghua, LI Baihui, WU Zexin, FANG Bo, LI Wei. Effects of myricetin on proliferation and apoptosis of glioma GL261 cells in mice [J]. Journal of Jilin University(Medicine Edition), 2018, 44(05): 955-961. |
| [15] | LIANG Bing, LIU Yang, SONG Jinping, QU Danhua, LIU Chunyan. Enhancement of histone deacetylase inhibitors SAHA in chemotherapeutic sensitivity of ovarian cancer SKOV3 cells [J]. Journal of Jilin University Medicine Edition, 2018, 44(03): 471-476. |
|
||