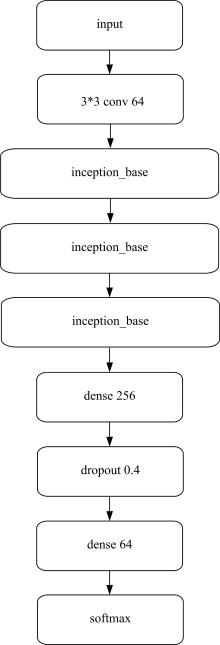
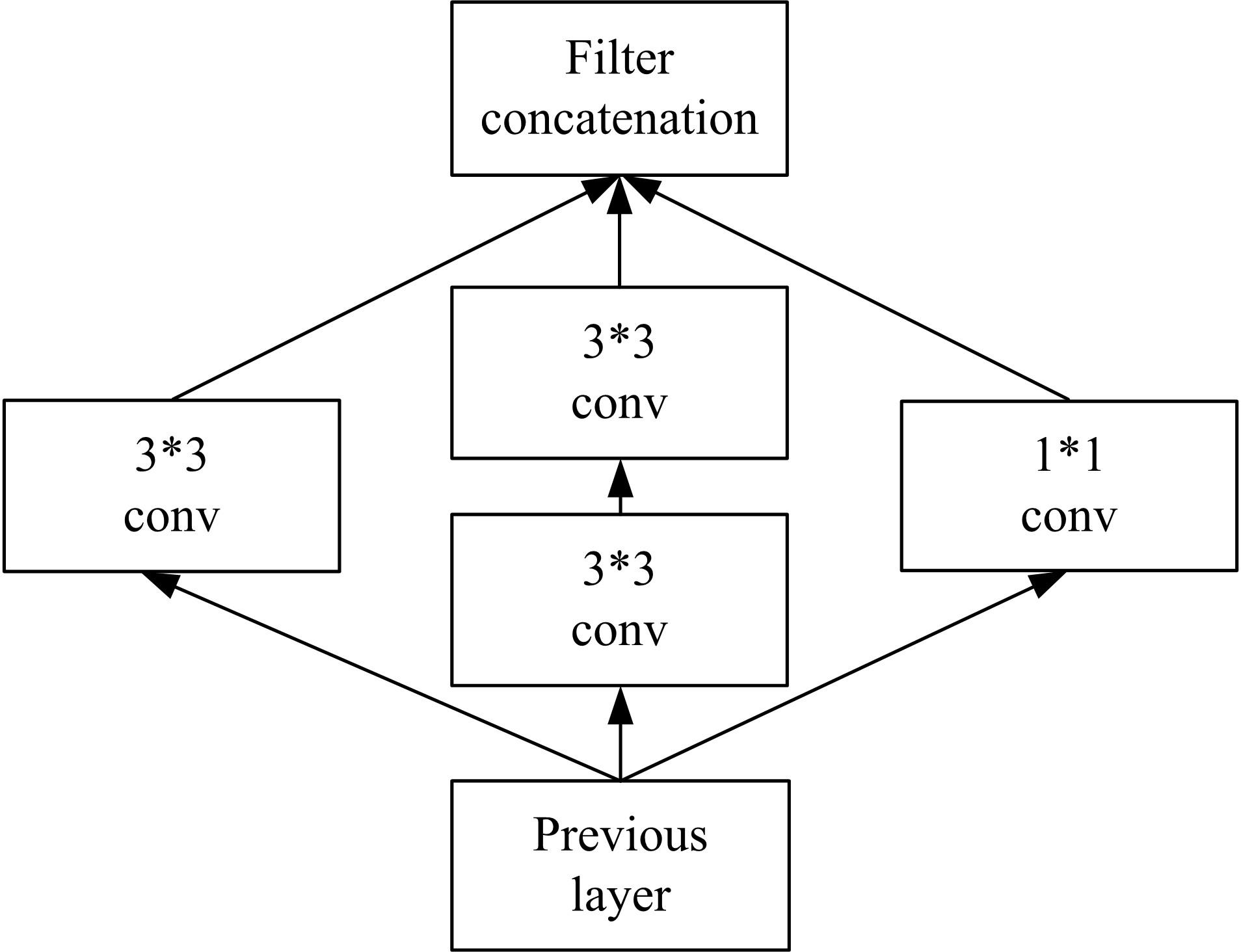
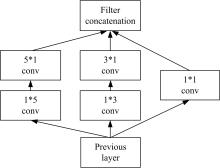
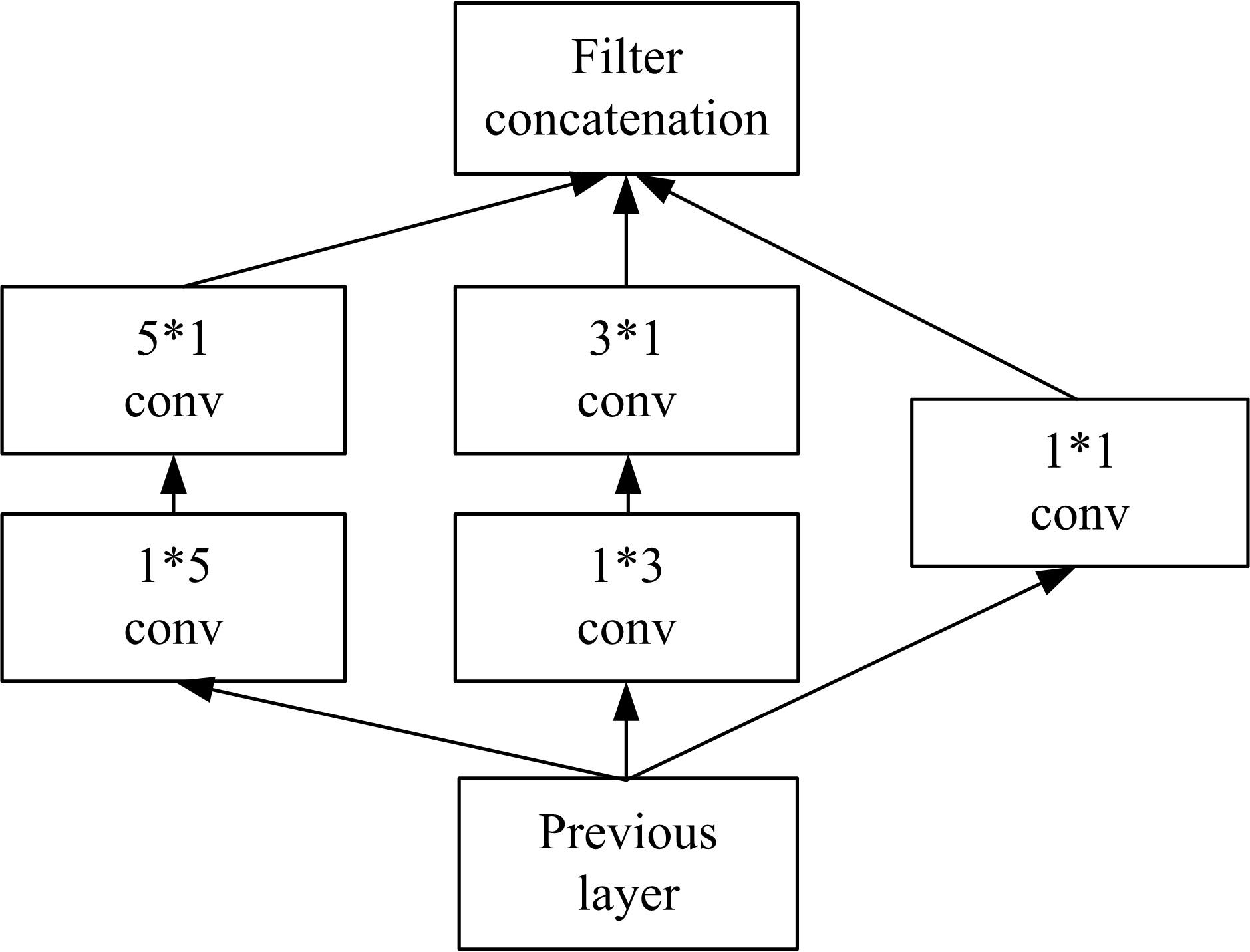
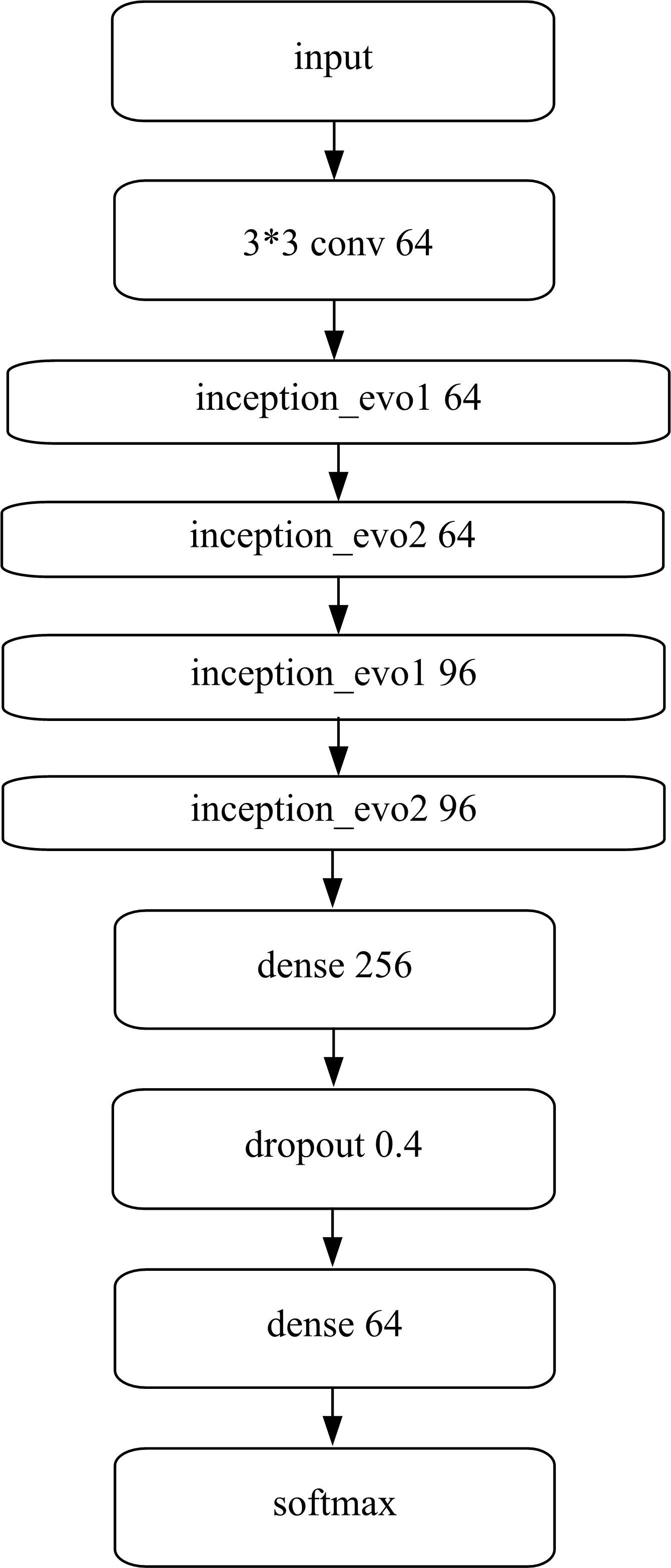
Journal of Jilin University(Engineering and Technology Edition) ›› 2022, Vol. 52 ›› Issue (1): 187-194.doi: 10.13229/j.cnki.jdxbgxb20200723
Prediction of protein-ATP binding site based on deep learning
Gui-xia LIU1,2( ),Zhi-yao PEI1,2,Jia-zhi SONG1,2
),Zhi-yao PEI1,2,Jia-zhi SONG1,2
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbol Computation and Knowledge Engineering of Ministry Education,Jilin University,Changchun 130012,China
CLC Number:
- TP399
| 1 | Bain F E, Fischer L A, Chen R, et al. Chapter seventeen-single-molecule analysis of replication protein A⁃DNA interactions[J]. Methods in Enzymology, 2018, 600: 439-461. |
| 2 | Raphael T, Timo M, Schroda M, et al. ATP-dependent molecular chaperones in plastids — more complex than expected[J]. Biochimica et Biophysica Acta Bioenergetics, 2015, 1847(9):872-888. |
| 3 | Gerwert K, Freier E, Wolf S. The role of protein-bound water molecules in microbial rhodopsins[J]. Biochimica Et Biophysica Acta, 2014, 1837(5): 606-613. |
| 4 | Qi W, Zhen L P, Yang Z, et al. COACH-D: improved protein⁃ligand binding sites prediction with refined ligand-binding poses through molecular docking[J]. Nuclc Acids Research,2018,46(W1):438-442. |
| 5 | Baldassi C, Zamparo M, Feinauer C, et al. Fast and accurate multivariate gaussian modeling of protein families: predicting residue contacts and protein-interaction partners[J]. PLoS ONE, 2014, 9(3): No. e92721. |
| 6 | 冉军舰. 赵瑞香 . 梁新红 ,等.基于蛋白互作网络解析甘薯渣中去氢表雄酮的生物作用机制[J]. 扬州大学学报:农业与生命科学版,2019, 40(5): 33-38. |
| Ran Jun-jian, Zhao Rui-xiang, Liang Xin-hong, et al. Biological mechanism analysis of DHEA from sweet potato residue based on protein-protein interaction network[J]. Journal of Yangzhou University: Agricultural and Life Science Edition, 2019, 40(5): 33-38. | |
| 7 | Carrano A, Snkhchyan H, Kooij G, et al. ATP-binding cassette transporters P-glycoprotein and breast cancer related protein are reduced in capillary cerebral amyloid angiopathy[J]. Neurobiology of Aging, 2014, 35(3): 565-575. |
| 8 | Christian S, Christoph W, Walter M. ATP- and ADP-dnaA protein, a molecular switch in gene regulation[J]. Embo Journal, 1999, 18(21): 6169-6176. |
| 9 | Chauhan J S, Mishra N K, Raghava G P. Identification of ATP binding residues of a protein from its primary sequence[J]. BMC Bioinformatics, 2009, 10(1): 434. |
| 10 | Chen K, Mizianty M J, Kurgan L. ATPsite: sequence-based prediction of ATP-binding residues[J]. Proteome Science, 2011, 9(S1):235-242. |
| 11 | Kurgan L, Chen K, Mizianty M J. Prediction and analysis of nucleotide binding residues using sequence and sequence-derived structural descriptors[J]. Bioinformatics, 2011, 28(3):331-341. |
| 12 | Yu Dong-Jun, Hu jun, Tang Zhen-min, et al. Improving protein-ATP binding residues prediction by boosting SVMs with random under-sampling[J]. Neurocomputing, 2013, 104: 180-190. |
| 13 | Yu Dong-jun, Hu Jun, Huang Yan, et al. TargetATPsite: a template‐free method for ATP‐binding sites prediction with residue evolution image sparse representation and classifier ensemble[J]. Journal of Computational Chemistry, 2013, 34(11): 974-985. |
| 14 | Fang C, Noguchi T, Yamana H. Simplified sequence-based method for ATP-binding prediction using contextual local evolutionary conservation[J]. Algorithms for Molecular Biology, 2014, 9(1): 7. |
| 15 | Andrews B J, Hu J. TSC_ATP: a two-stage classifier for predicting protein-ATP binding sites from protein sequence[C]∥ Computational Intelligence in Bioinformatics & Computational Biology, Niagara Falls, ON, Canada, 2015: 1-5. |
| 16 | Hu Jun, Li Yang, Zhang Yang,et al. ATPbind: accurate protein⁃ATP binding site prediction by combining sequence-profiling and structure-based comparisons[J]. Journal of Chemical Information & Modeling, 2018,58:501-510. |
| 17 | Zhou J, Lu Q, Xu R, et al. CNNsite: prediction of DNA-binding residues in proteins using convolutional neural network with sequence features[C]∥IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 2016: 78-85. |
| 18 | Pan Xiao-yong, Peter R, Yan Jun-chi, et al. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks[J]. BMC Genomics, 2018, 19(1):511. |
| 19 | Zhang Yu, Yu Dong-jun. Protein-ATP binding site prediction based on 1D-convolutional neural network[J]. Journal of Computer Applications, 2019, 39(11): 3146-3150. |
| 20 | Mcguffin L J, Kevin B, Jones D T. The PSIPRED protein structure prediction server[J]. Bioinformatics, 2000,16(4): 2. |
| 21 | Faraggi E, Zhou Yao-qi, Kloczkowski A. Accurate single-sequence prediction of solvent accessible surface area using local and global features[J]. Protns Structure Function & Bioinformatics, 2015, 82(11): 3170-3176. |
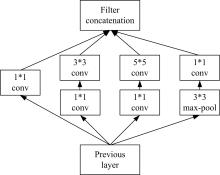
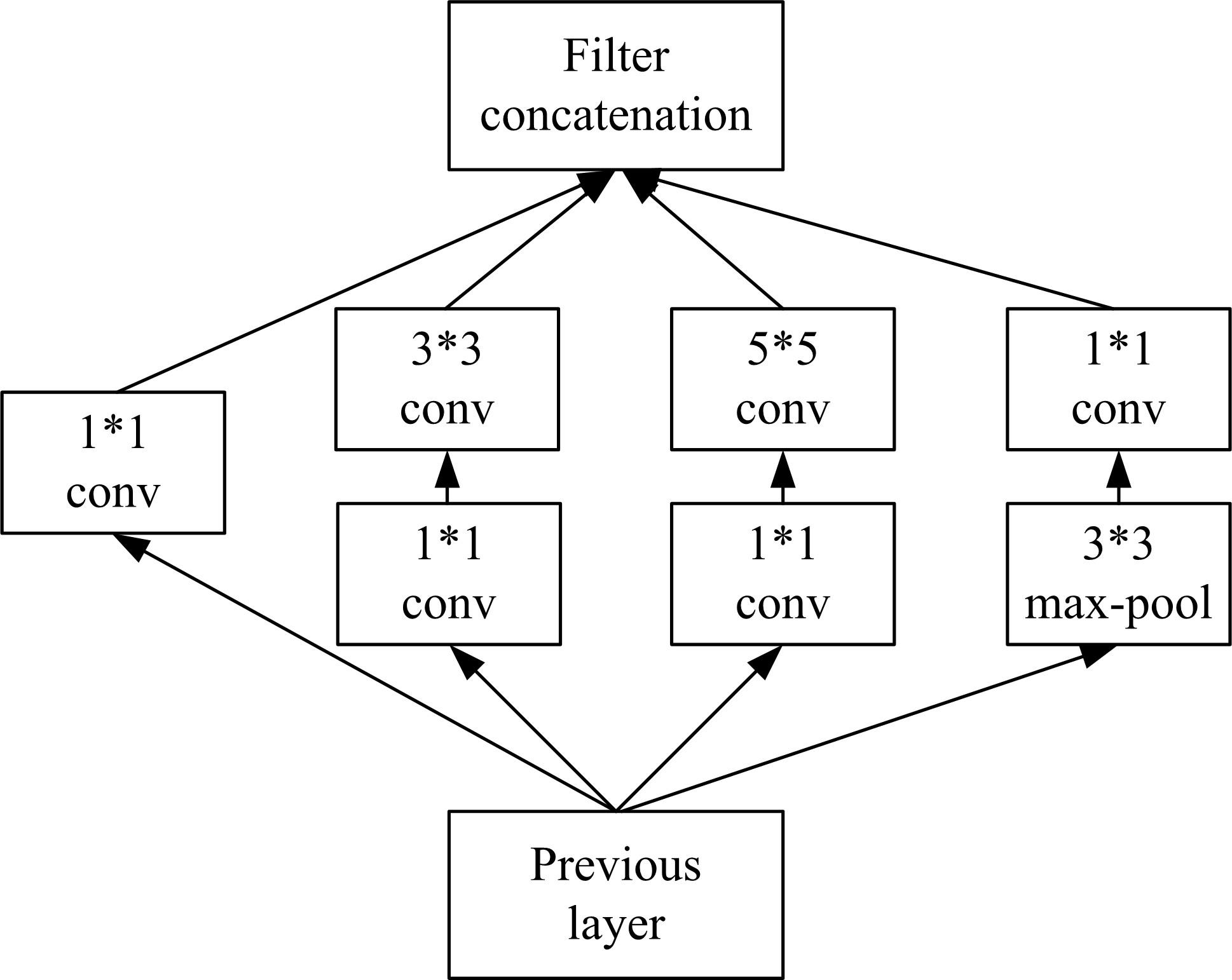
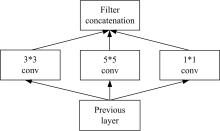
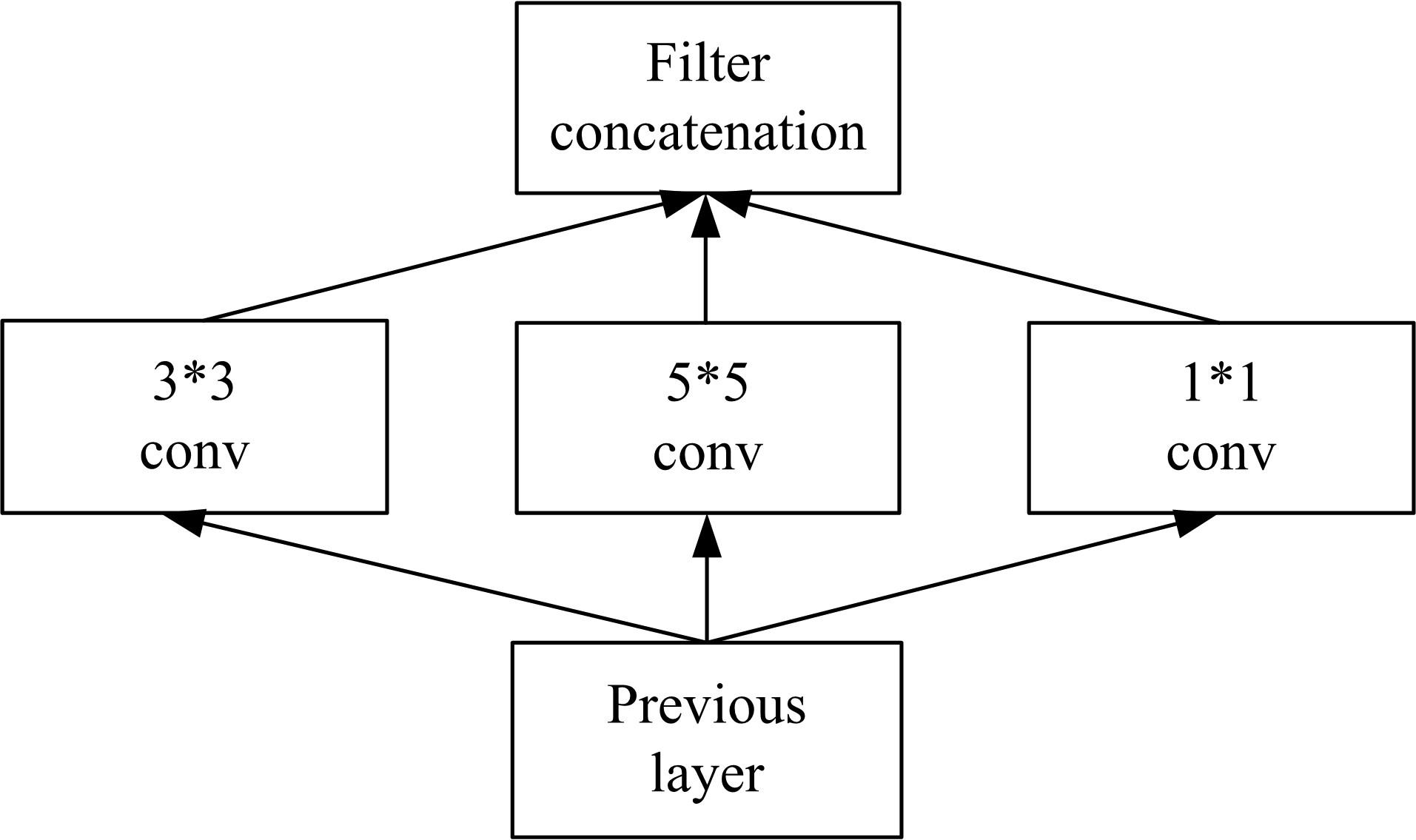
| 22 | Szegedy C, Liu W, Jia Y, et al. Going Deeper with Convolutions[C]∥ IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 2015:1-9. |
| 23 | Szegedy C, Ioffe S, Vanhoucke V, et al. Rethinking the inception architecture for computer vision[C]∥IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 2016:2818-2826. |
| 24 | Lin T Y, Goyal P, Girshick R, et al. Focal loss for dense object detection[J]. IEEE Transactions on Pattern Analysis & Machine Intelligence, 2017,42(2): 318-327. |
| 25 | Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning[J]. Computer Vision and Pattern Recognition,2016: arXiv:. |
| [1] | You QU,Wen-hui LI. Single-stage rotated object detection network based on anchor transformation [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(1): 162-173. |
| [2] | Jie ZHANG,Wen JING,Fu CHEN. Vulnerability detection of instant messaging network protocol based on passive clustering algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(6): 2253-2258. |
| [3] | Yuan-hong LIU,Pan-pan GUO,Yan-sheng ZHANG,Xin LI. Feature extraction of sparse graph preserving projection based on Riemannian manifold [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(6): 2268-2279. |
| [4] | Li-li DONG,Dan YANG,Xiang ZHANG. Large⁃scale semantic text overlapping region retrieval based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(5): 1817-1822. |
| [5] | Hui ZHONG,Heng KANG,Ying-da LYU,Zhen-jian LI,Hong LI,Ruo-chuan OUYANG. Image manipulation localization algorithm based on channel attention convolutional neural networks [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(5): 1838-1844. |
| [6] | Li-sheng JIN,Bai-cang GUO,Fang-rong WANG,Jian SHI. Dynamic multiple object detection algorithm for vehicle forward based on improved YOLOv3 [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(4): 1427-1436. |
| [7] | Xiao-long ZHU,Zhong XIE. Automatic construction of knowledge graph based on massive text data [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(4): 1358-1363. |
| [8] | Feng-chong LAN,Ji-wen LI,Ji-qing CHEN. DG-SLAM algorithm for dynamic scene compound deep learning and parallel computing [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(4): 1437-1446. |
| [9] | Jin-qing LI,Jian ZHOU,Xiao-qiang DI. Learning optical image encryption scheme based on CycleGAN [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(3): 1060-1066. |
| [10] | Zhe-ming YUAN,Hong-jie YUAN,Yu-xuan YAN,Qian LI,Shuang-qing LIU,Si-qiao TAN. Automatic recognition and classification of field insects based on lightweight deep learning model [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(3): 1131-1139. |
| [11] | Bo PENG,Yuan-yuan ZHANG,Yu-ting WANG,Ju TANG,Ji-ming XIE. Automatic traffic state recognition from videos based on auto⁃encoder and classifiers [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(3): 886-892. |
| [12] | Zhen SONG,Jun-liang LI,Gui-qiang LIU. Constant flow prediction method of variable speed hydraulic power source based on deep learning and limitation fuzzy [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(3): 1106-1110. |
| [13] | Tao XU,Ke MA,Cai-hua LIU. Multi object pedestrian tracking based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2021, 51(1): 27-38. |
| [14] | Hong-wei ZHAO,Xiao-han LIU,Yuan ZHANG,Li-li FAN,Man-li LONG,Xue-bai ZANG. Clothing classification algorithm based on landmark attention and channel attention [J]. Journal of Jilin University(Engineering and Technology Edition), 2020, 50(5): 1765-1770. |
| [15] | Hua CHEN,Wei GUO,Jing-wen YAN,Wen-hao ZHUO,Liang-bin WU. A new deep learning method for roads recognition from SAR images [J]. Journal of Jilin University(Engineering and Technology Edition), 2020, 50(5): 1778-1787. |
|
||