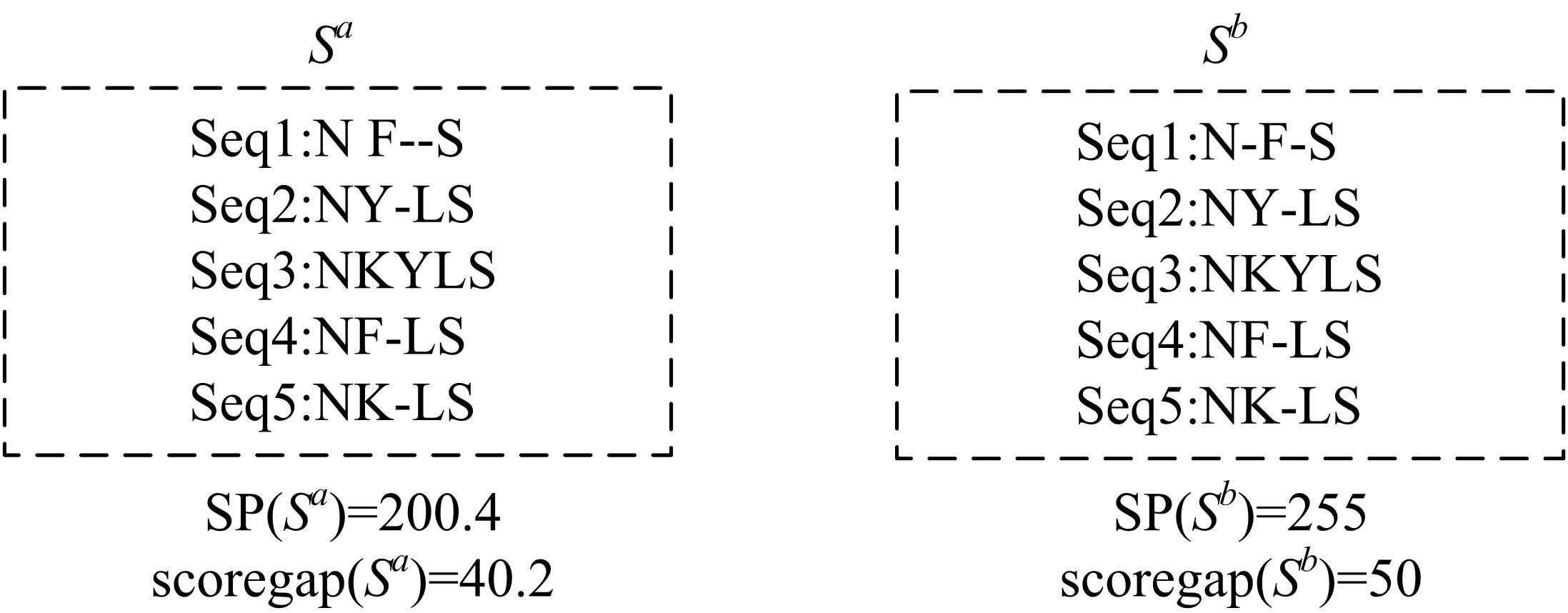
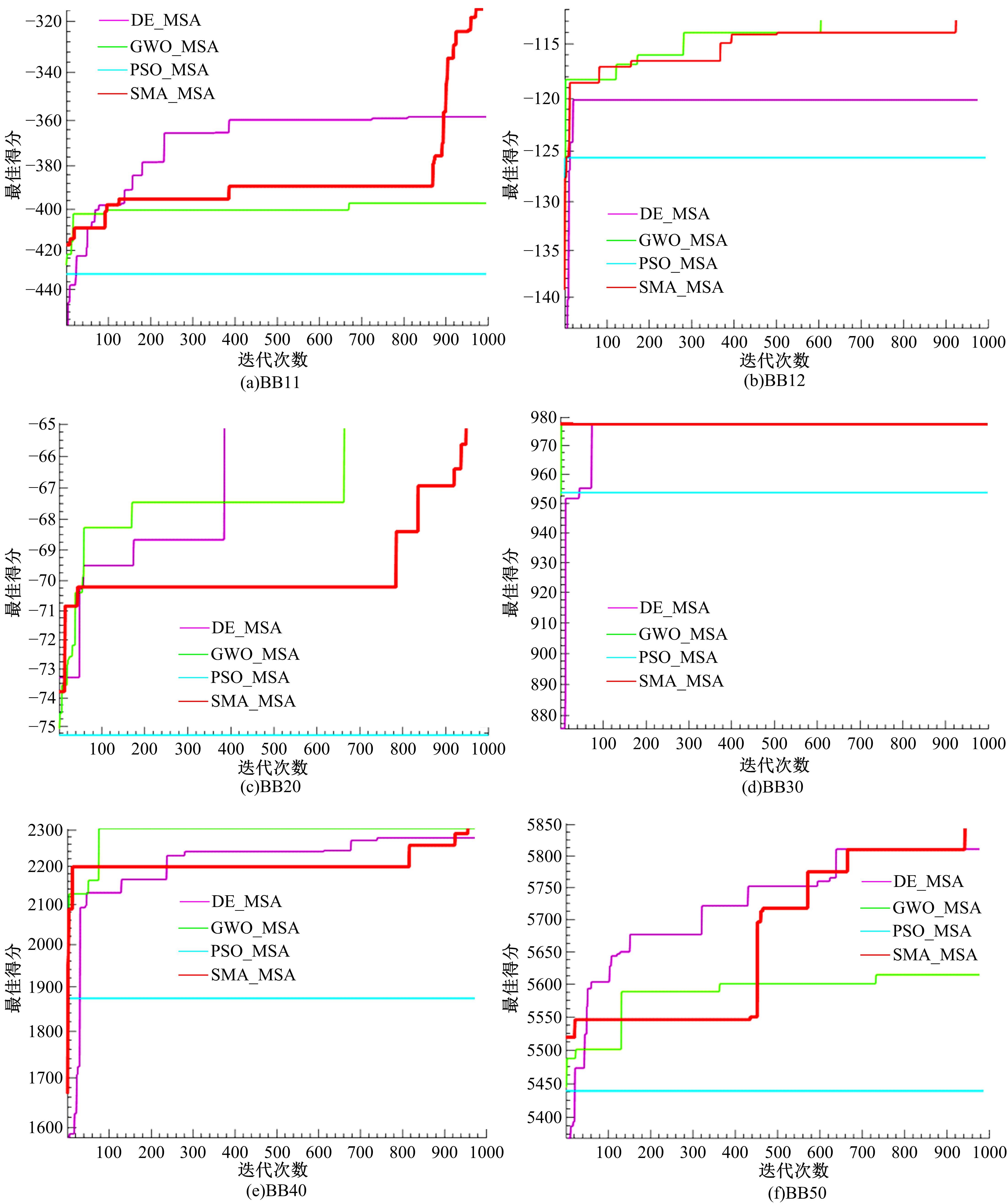
Journal of Jilin University(Engineering and Technology Edition) ›› 2022, Vol. 52 ›› Issue (12): 2984-2993.doi: 10.13229/j.cnki.jdxbgxb20210538
Previous Articles Next Articles
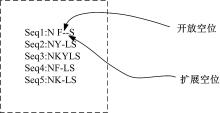
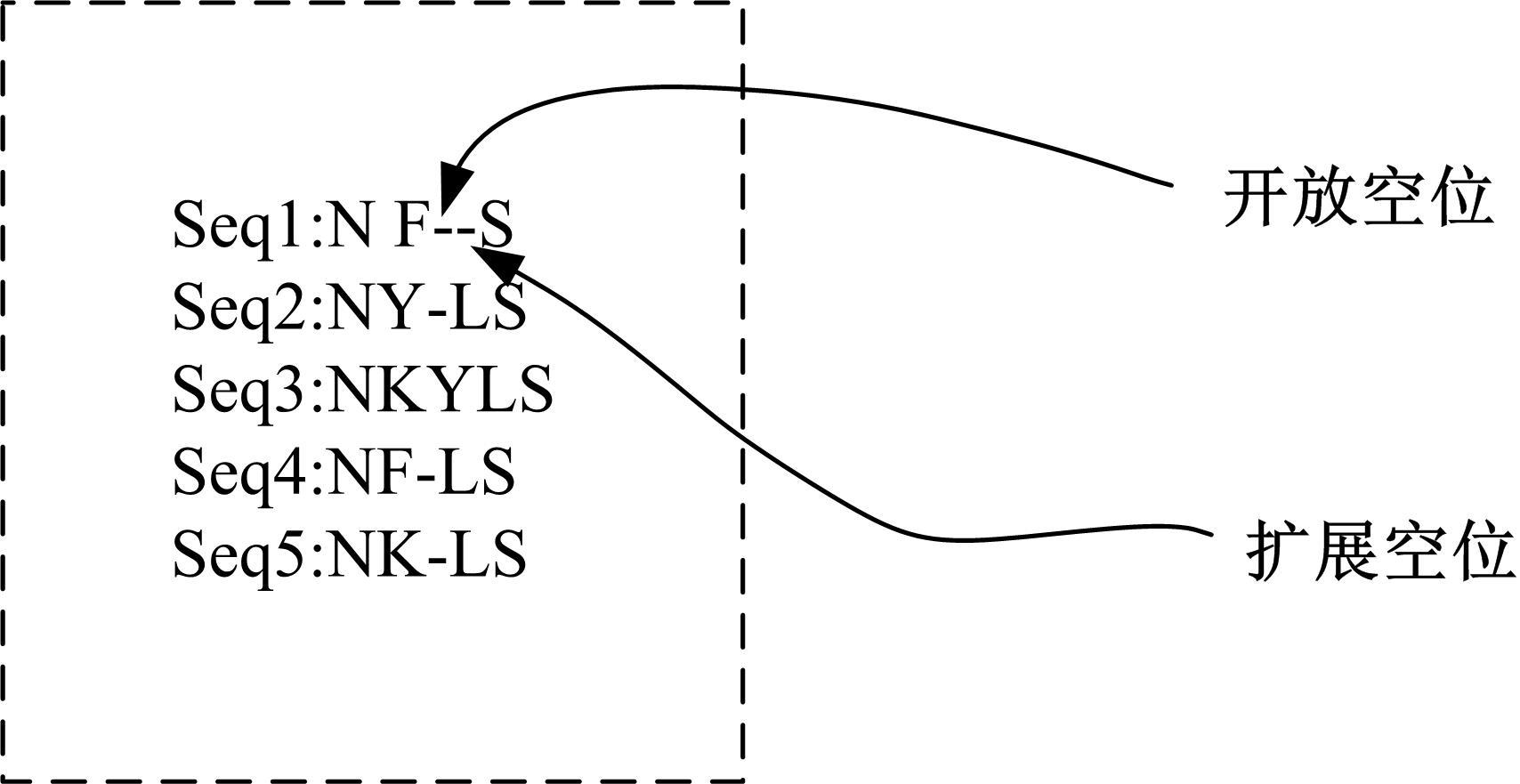
Multiple sequence alignment of proteins based on slime mold algorithm
Xin-lu WANG1,2,3( ),Da-you LIU1,2(
),Da-you LIU1,2( ),Si-han LIU1,3,Zheng WANG1,2,Li-wei ZHANG1,2,Sa DONG1,2
),Si-han LIU1,3,Zheng WANG1,2,Li-wei ZHANG1,2,Sa DONG1,2
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbolic Computation and Knowledge Engineering,Ministry of Education,Jilin University,Changchun 130012,China
3.College of International Education,Jilin University,Changchun 130012,China
CLC Number:
- TP391
| 1 | Eric S, Martinez H M. A multiple sequence alignment program[J]. Nucleic Acids Research, 1986(1):363-374. |
| 2 | Layeb A, Boudra A, Korichi W, et al. A new greedy randomized adaptive search procedure for multiobjective RNA structural alignment[J]. International Journal in Foundations of Computer Science & Technology, 2013, 3(1): 1-14. |
| 3 | 李文. 基于k-mer相异度算法在系统进化关系中的应用[D]. 广州:华南理工大学物理与光电学院, 2019. |
| Li Wen. Application of dissimilarity algorithms based on k-mer in evolutionary relationship[D]. Guangzhou: School of Physics, South China University of Technology, 2019. | |
| 4 | Edgar R C. Muscle: a multiple sequence alignment method with reduced time and space complexity[J]. BMC Bioinformatics, 2004, 5(1): No.113. |
| 5 | Lassmann T, Sonnhammer E L. Kalign—an accurate and fast multiple sequence alignment algorithm[J]. BMC Bioinformatics, 2005, 6(1): 1-9. |
| 6 | Paolo D T, Sebastien M, Ioannis X, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension[J]. Nucleic Acids Research, 2011, 39: W13-W17. |
| 7 | Wallace I M, Orla O, Higgins D G, et al. M-Coffee: combining multiple sequence alignment methods with T-Coffee[J]. Nucleic Acids Research, 2006, 34(6): 1692-1699. |
| 8 | Katoh K, Kuma K I, Miyata T, et al. Improvement in the accuracy of multiple sequence alignment program MAFFT[J]. Genome Informatics, 2005, 16(1):22-33. |
| 9 | Notredame C.Recent Evolutions of multiple sequence alignment algorithms[J]. Plos Computational Biology, 2007, 3: No.e123. |
| 10 | Ling C, Wei L, Chen J. Ant colony optimization methodfor multiple sequence alignment[C]∥International Conference on Machine Learning & Cybernetics, Piscatanay, NJ, 2007: 914-919. |
| 11 | Rubio-Largo A, Vega-Rodríguez M A, González-Álvarez D L. A hybrid multiobjective memetic metaheuristic for multiple sequence alignment[J]. IEEE Transactions on Evolutionary Computation, 2016, 20(4): 499-514. |
| 12 | Liu Y, Schmidt B, Maskell D L. Msaprobs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities[J]. Bioinformatics, 2010, 26: 1958-1964. |
| 13 | Gao Y X. A multiple sequence alignment algorithm based on inertia weights particle swarm optimization[J]. Journal of Bionanoence, 2014, 8(5): 400-404. |
| 14 | Rani R R, Ramyachitra D. Multiple sequence alignment using multi-objective based bacterial foraging optimization algorithm[J]. Biosystems, 2016, 150: 177-189. |
| 15 | Sun J, Wu X, Wei F, et al. Multiple sequence alignment using the hidden markov model trained by an improved quantum-behaved particle swarm optimization[J]. Information Sciences, 2012, 182(1): 93-114. |
| 16 | Öztürk C, Aslan S. A new artificial bee colony algorithm to solve the multiple sequence alignment problem[J]. International Journal of Data Mining & Bioinformatics, 2016, 14(4): 332-353. |
| 17 | Zhu H, He Z, Jia Y. A novel approach to multiple sequence alignment using multiobjective evolutionary algorithm based on decomposition[J]. IEEE Journal of Biomedical & Health Informatics, 2016, 20(2): No.717. |
| 18 | Zambrano-Vega C, Nebro A J, Durillo J, et al. Multiple sequence alignment with multiobjective metaheuristics. a comparative study[J]. International Journal of Intelligent Systems, 2017, 32(8): 843-861. |
| 19 | Mokaddem A, Hadj A B, Elloumi M. Refin-align: new refinement algorithm for multiple sequence alignment[J]. Informatica, 2019, 43(4):527-534. |
| 20 | Bonizzoni P, Vedova G D. The complexity of multiple sequence alignment with SP-score that is a metric[J]. Theoretical Computer Science, 2001, 259(1/2):63-79. |
| 21 | Needleman S B, Wunsch C D. A general method applicable to search for similarities in amino acid sequence of 2 proteins[J]. Journal of Molecular Biology, 1970, 48(3): 443-453. |
| 22 | Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice[J]. Nucleic Acids Research, 1994, 22(22): 1673-1680. |
| 23 | Notredame C, Holm L, Higgins D G. Coffee: an objective function for multiple sequence alignments[J]. Bioinformatics, 1998(5): 407-422. |
| 24 | Notredame C, Higgins D G, Heringa J . et al. T-Coffee: a novel method for fast and accurate multiple sequence alignment[J]. Journal of Molecular Biology, 2000, 302(1): 205-217. |
| 25 | O'Sullivan O, Karsten S, Chantal A, et al. 3Dcoffee: combining protein sequences and structures within multiple sequence alignments[J]. Journal of molecular biology, 2004, 340(2): 385-395. |
| 26 | Naznin F, Sarker R, Essam D. Progressive alignment method using genetic algorithm for multiple sequence alignment[J]. IEEE Transactions on Evolutionary Computation, 2012, 16: 615-631. |
| 27 | Naznin F, Sarker R, Essam D. Vertical decomposition with genetic algorithm for multiple sequence alignment[J]. BMC Bioinformatics, 2011, 12: No. 353. |
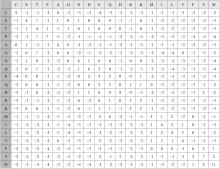
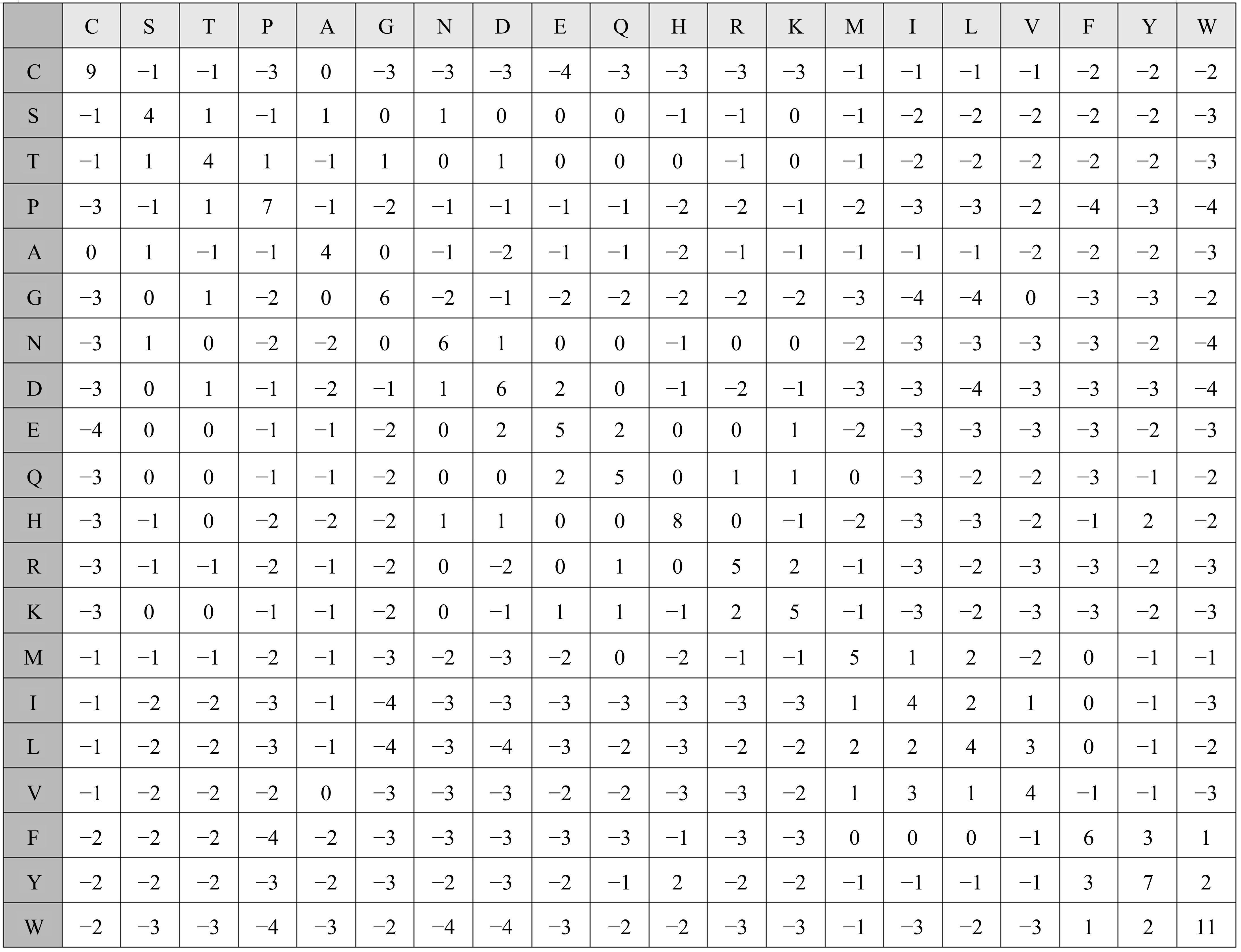
| 28 | Henikoff H. Amino acid substitution matrices from protein blocks[J]. Proceedings of the National Academy of Sciences, 1992, 89(22):10915-10919. |
| 29 | Li S, Chen H, Wang M, et al. Slime mould algorithm: a new method for stochastic optimization[J]. Future Generation Computer Systems, 2020, 111: 300-323. |
| 30 | Venter G, Jaroslaw S S. Particle swarm optimization[J]. AIAA Journal, 2003, 41(8): 129-132. |
| 31 | Mirjalili S, Mirjalili S, Lewis A. Grey wolf optimizer[J]. Advances in Engineering Software, 2014, 69: 46-61. |
| 32 | Das S S P N. Differential evolution: a survey of the state-of-the-art[J]. IEEE Transactions on Evolutionary Computation, 2011, 15(1): 4-31. |
| 33 | Thompson J D, Koehl P, Ripp R, et al. Balibase 3.0: latest developments of the multiple sequence alignment benchmark.[J]. Proteins-structure Function & Bioinformatics, 2010, 61(1): 127-136. |
| [1] | Xian-yu QI,Wei WANG,Lin WANG,Yu-fei ZHAO,Yan-peng DONG. Semantic topological map building with object semantic grid map [J]. Journal of Jilin University(Engineering and Technology Edition), 2023, 53(2): 569-575. |
| [2] | Xiao-hu SHI,Jia-qi WU,Chun-guo WU,Shi CHENG,Xiao-hui WENG,Zhi-yong CHANG. Residual network based curve enhanced lane detection method [J]. Journal of Jilin University(Engineering and Technology Edition), 2023, 53(2): 584-592. |
| [3] | Peng GUO,Wen-chao ZHAO,Kun LEI. Dual⁃resource constrained flexible job shop optimal scheduling based on an improved Jaya algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2023, 53(2): 480-487. |
| [4] | Jin-Zhen Liu,Guo-Hui Gao,Hui Xiong. Multi⁃scale attention network for brain tissue segmentation [J]. Journal of Jilin University(Engineering and Technology Edition), 2023, 53(2): 576-583. |
| [5] | Gui-he QIN,Jun-feng HUANG,Ming-hui SUN. Text input based on two⁃handed keyboard in virtual environment [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(8): 1881-1888. |
| [6] | Fu-heng QU,Tian-yu DING,Yang LU,Yong YANG,Ya-ting HU. Fast image codeword search algorithm based on neighborhood similarity [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(8): 1865-1871. |
| [7] | Tian BAI,Ming-wei XU,Si-ming LIU,Ji-an ZHANG,Zhe WANG. Dispute focus identification of pleading text based on deep neural network [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(8): 1872-1880. |
| [8] | Feng-feng ZHOU,Hai-yang ZHU. SEE: sense EEG⁃based emotion algorithm via three⁃step feature selection strategy [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(8): 1834-1841. |
| [9] | Feng-feng ZHOU,Yi-chi ZHANG. Unsupervised feature engineering algorithm BioSAE based on sparse autoencoder [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(7): 1645-1656. |
| [10] | Jun WANG,Yan-hui XU,Li LI. Data fusion privacy protection method with low energy consumption and integrity verification [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(7): 1657-1665. |
| [11] | Yao-long KANG,Li-lu FENG,Jing-an ZHANG,Fu CHEN. Outlier mining algorithm for high dimensional categorical data streams based on spectral clustering [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(6): 1422-1427. |
| [12] | Wen-jun WANG,Yin-feng YU. Automatic completion algorithm for missing links in nowledge graph considering data sparsity [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(6): 1428-1433. |
| [13] | Xue-yun CHEN,Xue-yu BEI,Qu YAO,Xin JIN. Pedestrian segmentation and detection in multi-scene based on G-UNet [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 925-933. |
| [14] | Shi-min FANG. Multiple source data selective integration algorithm based on frequent pattern tree [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 885-890. |
| [15] | Da-xiang LI,Meng-si CHEN,Ying LIU. Spontaneous micro-expression recognition based on STA-LSTM [J]. Journal of Jilin University(Engineering and Technology Edition), 2022, 52(4): 897-909. |
|