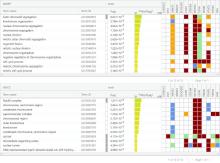
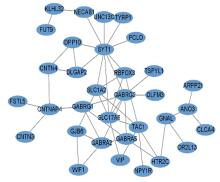
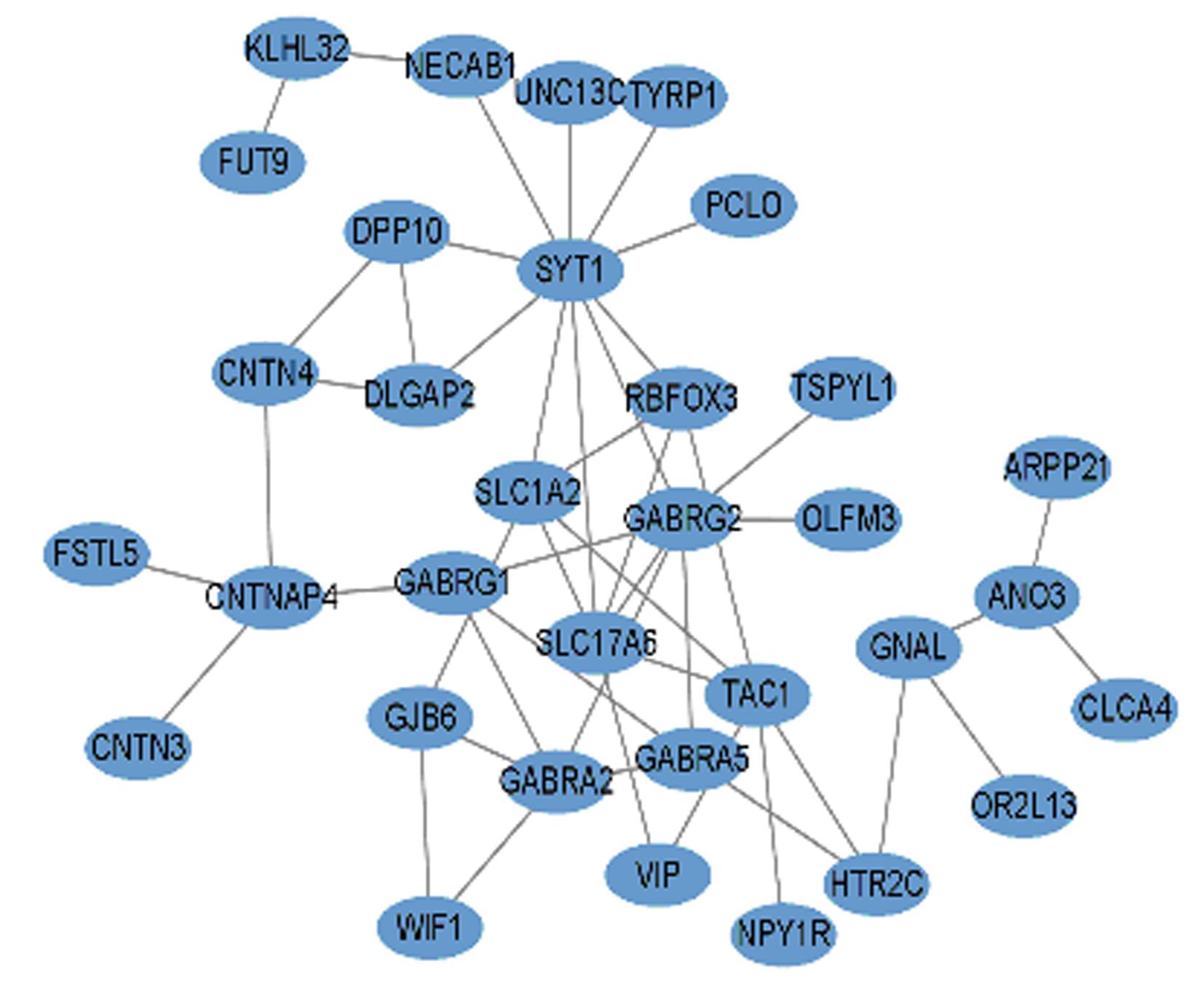
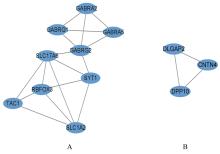
| 1 |
GOODENBERGER M L, JENKINS R B. Genetics of adult glioma[J]. Cancer Genet, 2012, 205(12): 613-621.
|
| 2 |
LOUIS D N, PERRY A, REIFENBERGER G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary[J]. Acta Neuropathol, 2016, 131(6): 803-820.
|
| 3 |
MØLLER H G, RASMUSSEN A P, ANDERSEN H H,et al. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion[J].Mol Neurobiol, 2013, 47(1): 131-144.
|
| 4 |
VERHAAK R G W, HOADLEY K A, PURDOM E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA,IDH1,EGFR,and NF1[J]. Cancer Cell, 2010, 17(1): 98-110.
|
| 5 |
CLAUS E B, WALSH K M, WIENCKE J K, et al. Survival and low-grade glioma: the emergence of genetic information[J]. Neurosurg Focus, 2015, 38(1): E6.
|
| 6 |
ALIFIERIS C, TRAFALIS D T. Glioblastoma multiforme: pathogenesis and treatment[J]. Pharmacol Ther, 2015, 152: 63-82.
|
| 7 |
THAKKAR J P, DOLECEK T A, HORBINSKI C, et al. Epidemiologic and molecular prognostic review of glioblastoma[J]. Cancer Epidemiol Biomarkers Prev, 2014, 23(10): 1985-1996.
|
| 8 |
MAHATA B, BISWAS S, RAYMAN P, et al. GBM derived gangliosides induce T cell apoptosis through activation of the caspase cascade involving both the extrinsic and the intrinsic pathway[J]. PLoS One, 2015, 10(7): e0134425.
|
| 9 |
MA H W, LI T, TAO Z N, et al. NKCC1 promotes EMT-like process in GBM via RhoA and Rac1 signaling pathways[J]. J Cell Physiol, 2019, 234(2): 1630-1642.
|
| 10 |
CHEN X J, LIN Q X, JIANG Y A, et al. Identification of potential biomarkers of platelet RNA in glioblastoma by bioinformatics analysis[J]. Biomed Res Int, 2022, 2022: 2488139.
|
| 11 |
REN Z J, HE Y, YANG Q Q, et al. A comprehensive analysis of the glutathione peroxidase 8 (GPX8) in human cancer[J]. Front Oncol, 2022, 12: 812811.
|
| 12 |
YUAN F, CAI X M, CONG Z X, et al. Roles of the m6A modification of RNA in the glioblastoma microenvironment as revealed by single-cell analyses[J]. Front Immunol, 2022, 13: 798583.
|
| 13 |
LIU J H, GAO L, JI B W, et al. BCL7A as a novel prognostic biomarker for glioma patients[J]. J Transl Med, 2021, 19(1): 335.
|
| 14 |
YU T L, ACHARYA A, MATTHEOS N, et al. Molecular mechanisms linking peri-implantitis and type 2 diabetes mellitus revealed by transcriptomic analysis[J]. PeerJ, 2019, 7: e7124.
|
| 15 |
PAOLACCI S, PRECONE V, ACQUAVIVA F,et al. Genetics of lipedema: new perspectives on genetic research and molecular diagnoses[J]. Eur Rev Med Pharmacol Sci, 2019, 23(13): 5581-5594.
|
| 16 |
OMURO A, DEANGELIS L M. Glioblastoma and other malignant gliomas: a clinical review[J]. JAMA, 2013, 310(17): 1842-1850.
|
| 17 |
SIEGHART W. Structure and pharmacology of gamma-aminobutyric acid A receptor subtypes[J]. Pharmacol Rev, 1995, 47(2): 181-234.
|
| 18 |
BLANCHART A, FERNANDO R, HÄRING M,et al. Endogenous GABAA receptor activity suppresses glioma growth[J]. Oncogene, 2017, 36(6): 777-786.
|
| 19 |
BARBIERI F, BOSIO A G, PATTAROZZI A, et al. Chloride intracellular channel 1 activity is not required for glioblastoma development but its inhibition dictates glioma stem cell responsivity to novel biguanide derivatives[J]. J Exp Clin Cancer Res, 2022,41(1): 53.
|
| 20 |
OLSEN R W, SIEGHART W. International Union of Pharmacology. LXX. Subtypes of gamma-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function Update[J]. Pharmacol Rev, 2008, 60(3): 243-260.
|
| 21 |
MAILLARD P Y, BAER S, SCHAEFER É, et al. Molecular and clinical descriptions of patients with GABAA receptor gene variants (GABRA1, GABRB2, GABRB3, GABRG2): a cohort study, review of literature, and genotype-phenotype correlation[J]. Epilepsia, 2022, 63(10): 2519-2533.
|
| 22 |
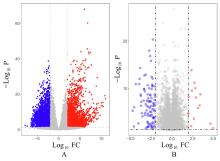
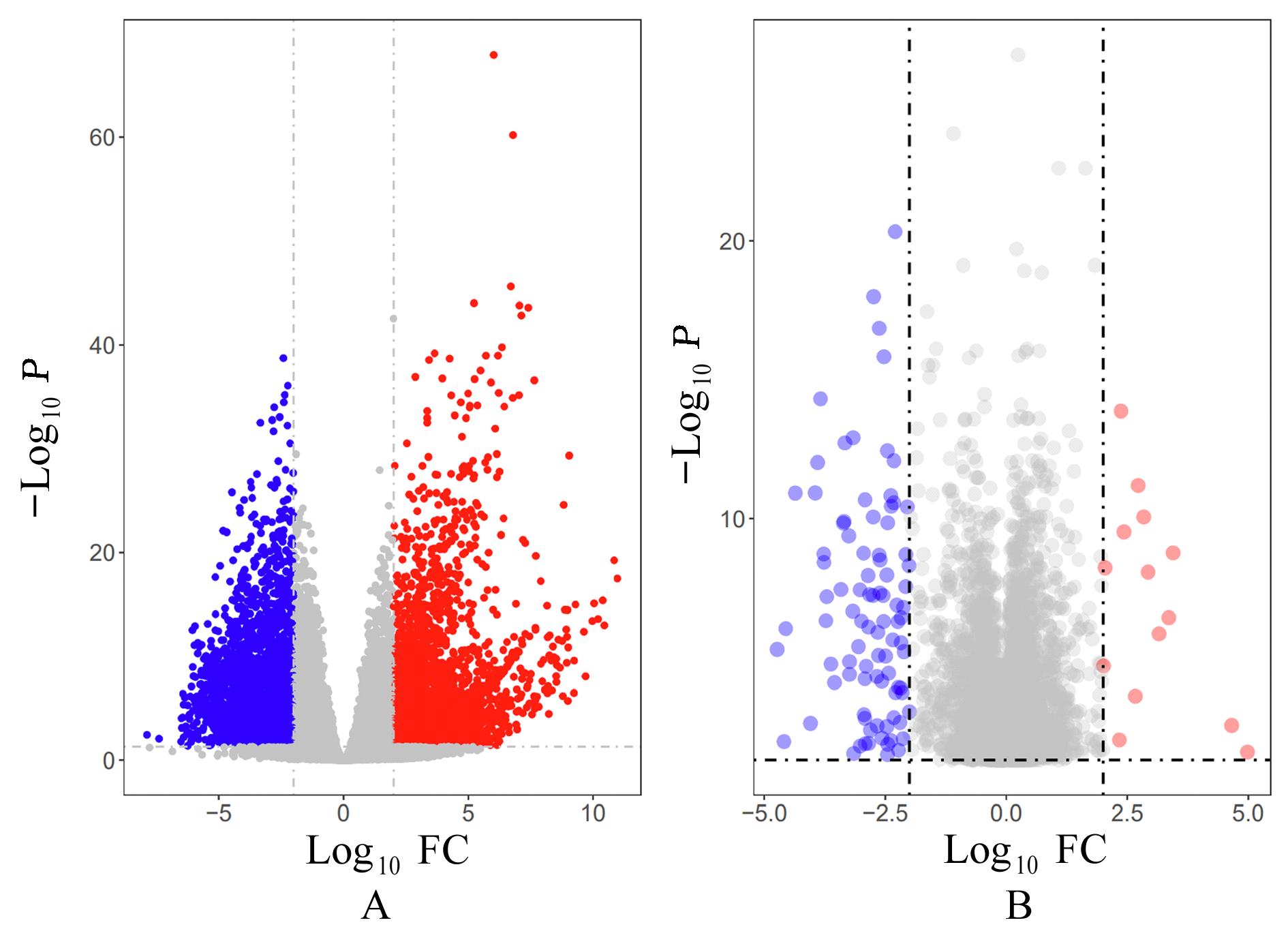
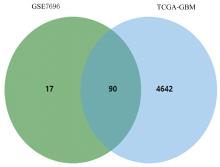
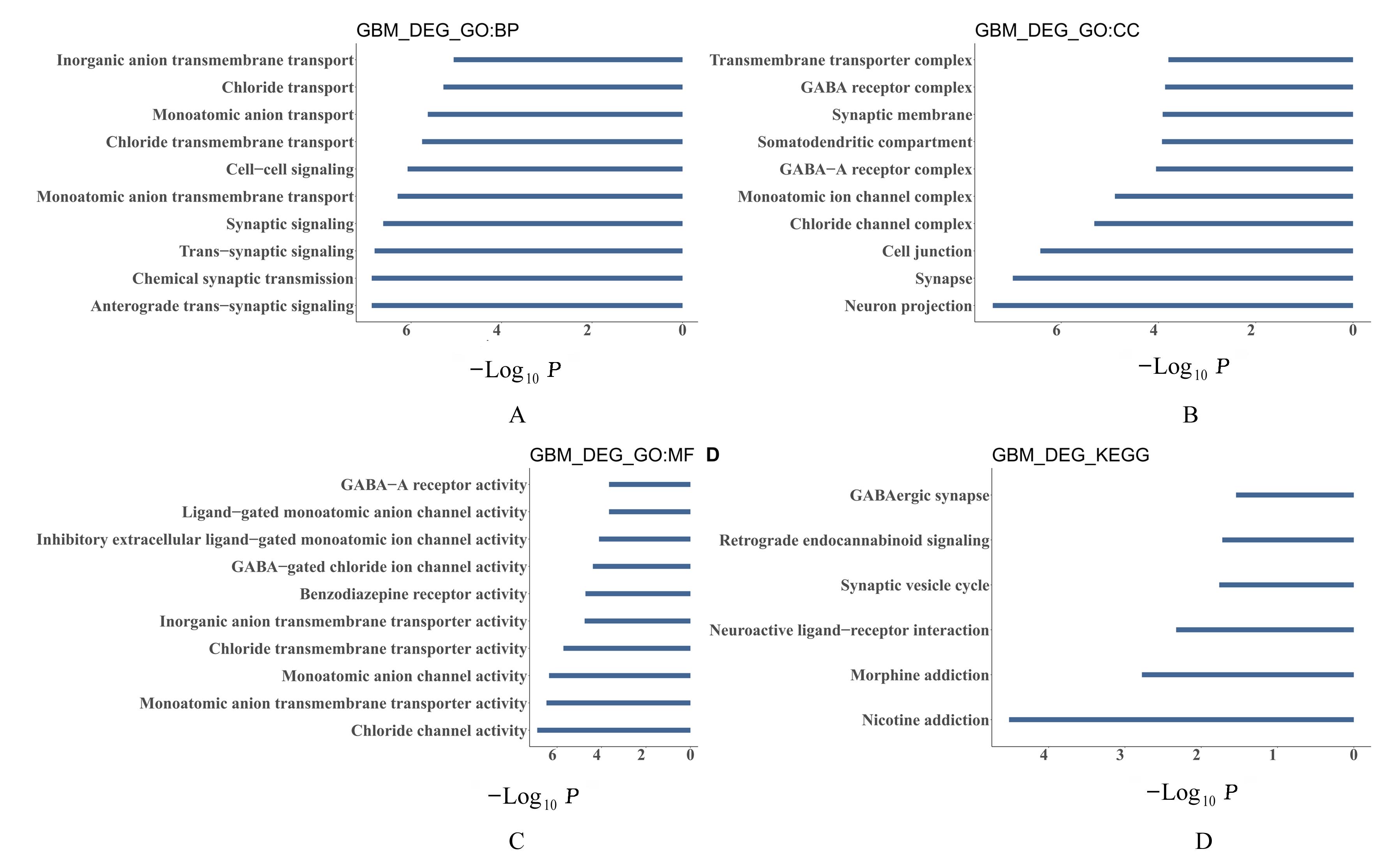
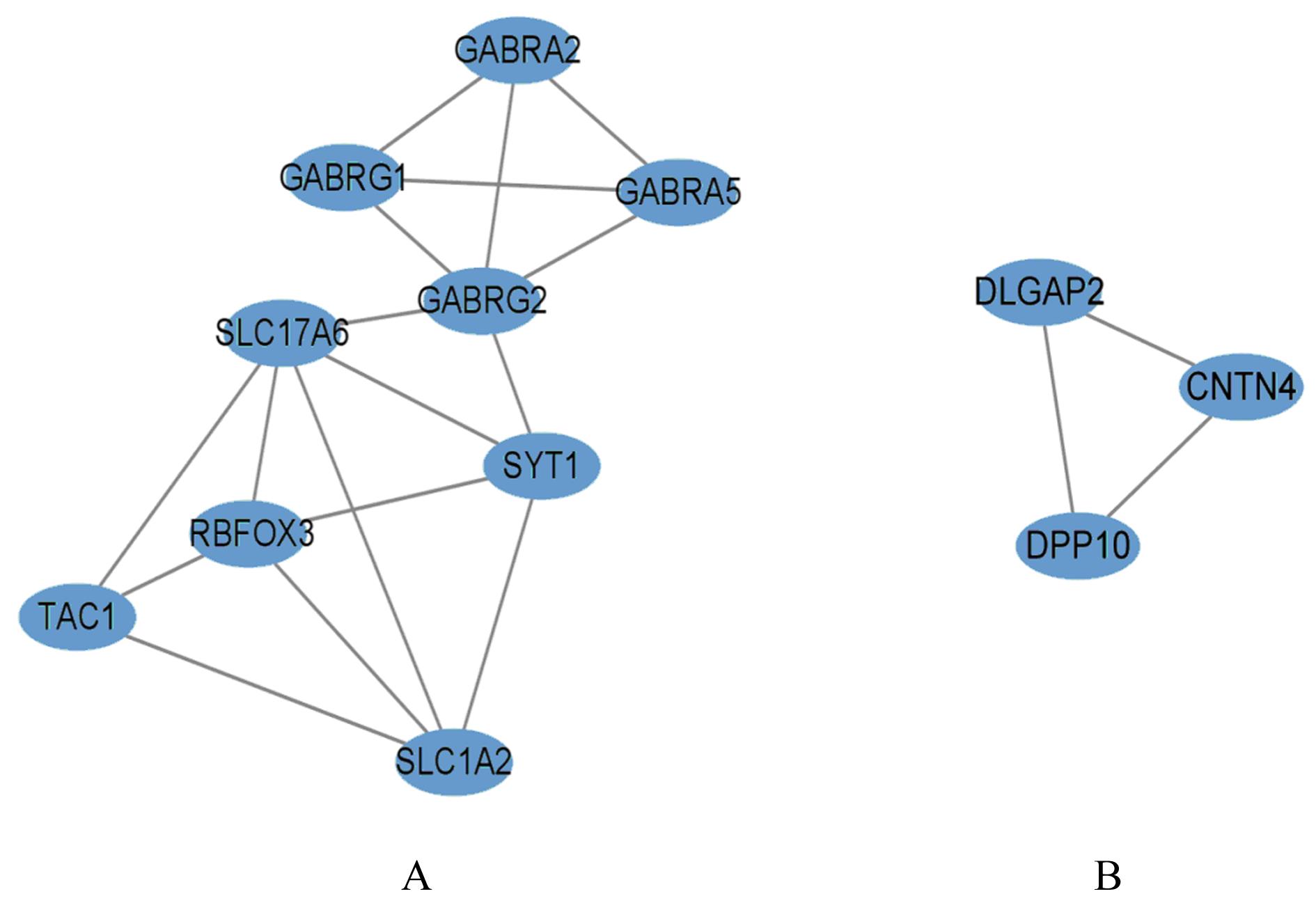
ZHANG X M, JIANG M Y, TANG S F, et al. Comprehensive bioinformatic analysis of key genes and signaling pathways in glioma[J].JUSTC, 2022, 52(9): 3.
|
| 23 |
SÜDHOF T C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle[J]. Neuron, 2013, 80(3): 675-690.
|
| 24 |
NORD H, HARTMANN C, ANDERSSON R, et al. Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array[J]. Neuro-oncology, 2009, 11(6): 803-818.
|
| 25 |
MISAWA K, KANAZAWA T, MISAWA Y, et al. Frequent promoter hypermethylation of tachykinin-1 and tachykinin receptor type 1 is a potential biomarker for head and neck cancer[J]. J Cancer Res Clin Oncol, 2013, 139(5): 879-889.
|
| 26 |
WRANGLE J, MACHIDA E O, DANILOVA L,et al. Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer[J]. Clin Cancer Res, 2014, 20(7): 1856-1864.
|
| 27 |
DUAN W, ZHANG Y P, HOU Z, et al. Novel insights into NeuN: from neuronal marker to splicing regulator[J]. Mol Neurobiol, 2016, 53(3): 1637-1647.
|
| 28 |
LIU T Z, LI W B, LU W J, et al. RBFOX3 promotes tumor growth and progression via hTERT signaling and predicts a poor prognosis in hepatocellular carcinoma[J]. Theranostics, 2017, 7(12): 3138-3154.
|
| 29 |
MARAGAKIS N J, ROTHSTEIN J D. Glutamate transporters: animal models to neurologic disease[J]. Neurobiol Dis, 2004, 15(3): 461-473.
|
| 30 |
CONSORTIUM E. De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies[J]. Am J Hum Genet, 2016, 99(2): 287-298.
|
| 31 |
YAO P S, KANG D Z, LIN R Y, et al. Glutamate/glutamine metabolism coupling between astrocytes and glioma cells: neuroprotection and inhibition of glioma growth[J]. Biochem Biophys Res Commun, 2014, 450(1): 295-299.
|