Journal of Jilin University(Medicine Edition) ›› 2023, Vol. 49 ›› Issue (5): 1243-1252.doi: 10.13481/j.1671-587X.20230518
• Research in clinical medicine • Previous Articles
Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis
Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI( ),Suzhen WANG(
),Suzhen WANG( )
)
- Department of Health Statistics,School of Public Health,Weifang Medical University,Weifang 261053,China
-
Received:2022-11-24Online:2023-09-28Published:2023-10-26 -
Contact:Fuyan SHI,Suzhen WANG E-mail:shifuyan@126.com;wangsz@wfmc.edu.cn
CLC Number:
- R735.7
Cite this article
Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis[J].Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252.
share this article
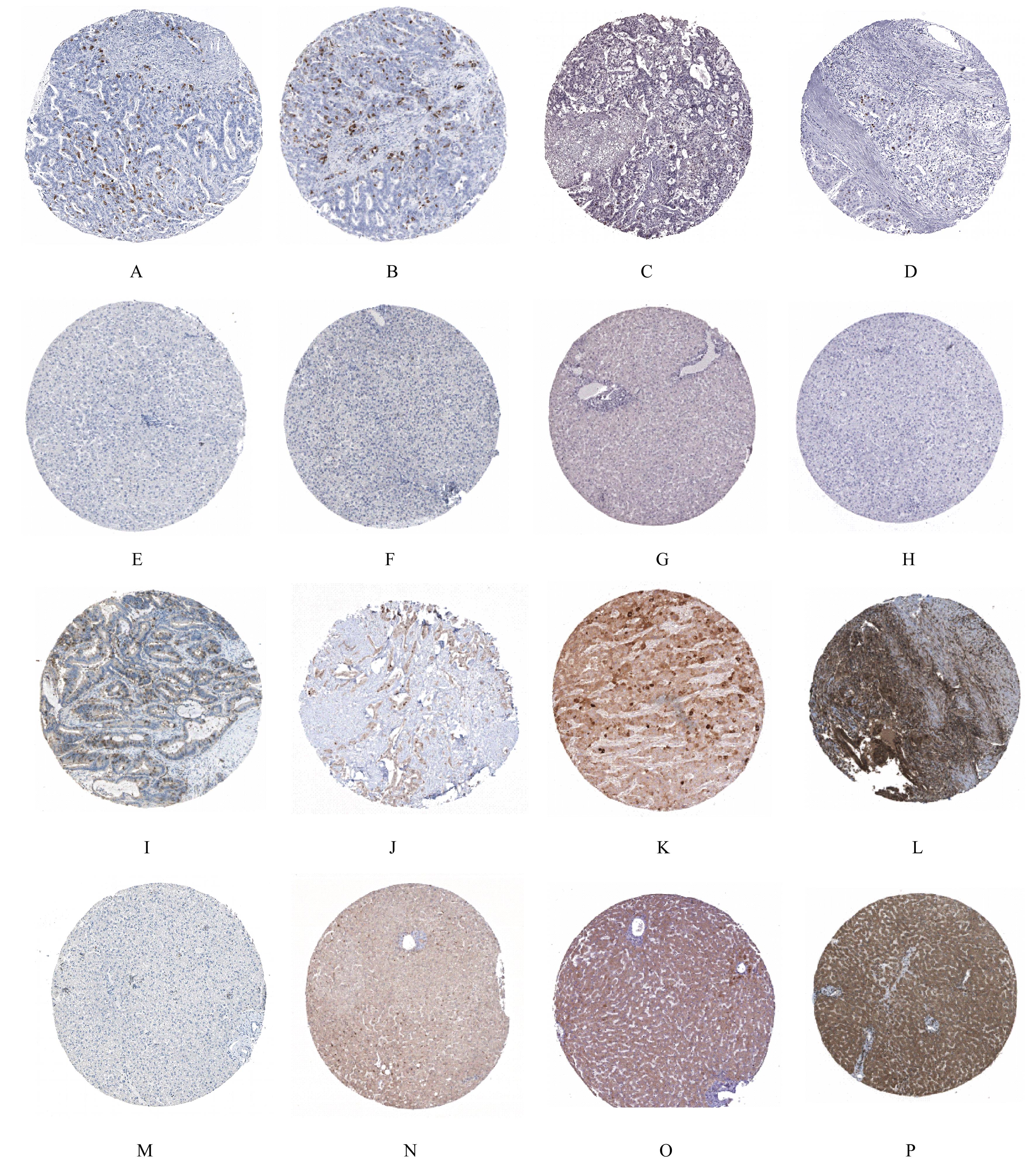
Tab. 1
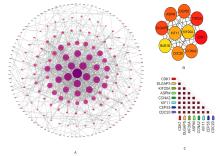
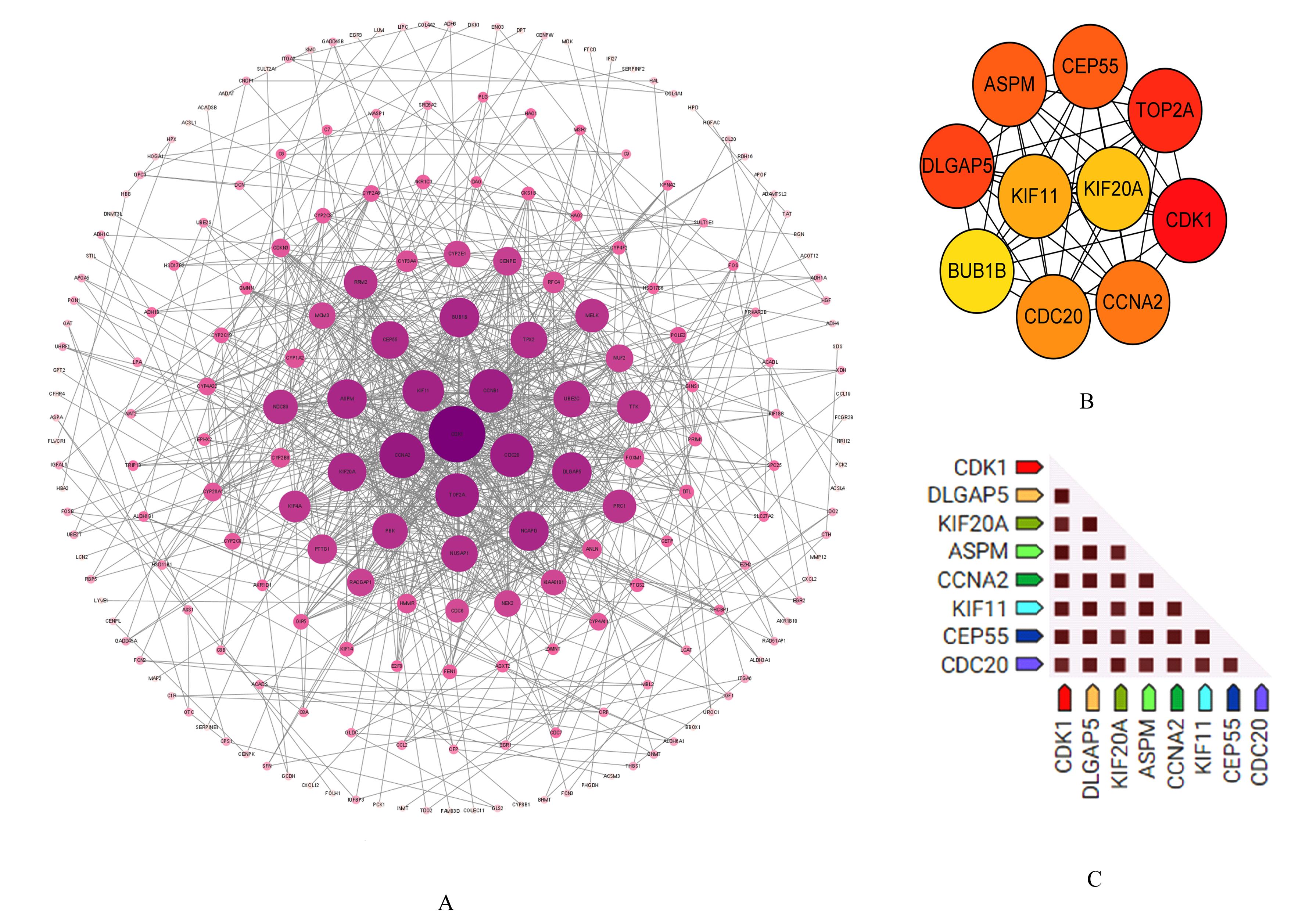
KEGG signaling pathway enrichment analysis on key genes"
| ID | Description | Count | P | Gene |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 4 | 2.702 3E-07 | CDC20,CDK1,CCNA2,BUB1B |
| hsa05203 | Viral carcinogenesis | 3 | 0.000 149 353 | CDC20,CDK1,CCNA2 |
| hsa05166 | Human T lymphocyte leukemia virus 1 infection | 3 | 0.000 192 061 | CDC20,CCNA2,BUB1B |
| hsa04914 | Progesterone-mediated oocyte maturation | 2 | 0.001 516 381 | CDK1,CCNA2 |
| hsa04114 | Oocyte meiosis | 2 | 0.002 488 770 | CDC20,CDK1 |
| hsa04218 | Cellular senescence | 2 | 0.003 511 844 | CDK1,CCNA2 |
| 1 | BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J].CA Cancer J Clin,2018,68(6):394-424. |
| 2 | PETRICK J L, FLORIO A A, ZNAOR A, et al. International trends in hepatocellular carcinoma incidence, 1978-2012[J]. Int J Cancer, 2020, 147(2): 317-330. |
| 3 | RINALDI L, GUARINO M, PERRELLA A, et al. Role of liver stiffness measurement in predicting HCC occurrence in direct-acting antivirals setting: a real-life experience[J]. Dig Dis Sci, 2019, 64(10): 3013-3019. |
| 4 | GRANDHI M S, KIM A K, RONNEKLEIV-KELLY S M, et al. Hepatocellular carcinoma: from diagnosis to treatment[J]. Surg Oncol, 2016, 25(2): 74-85. |
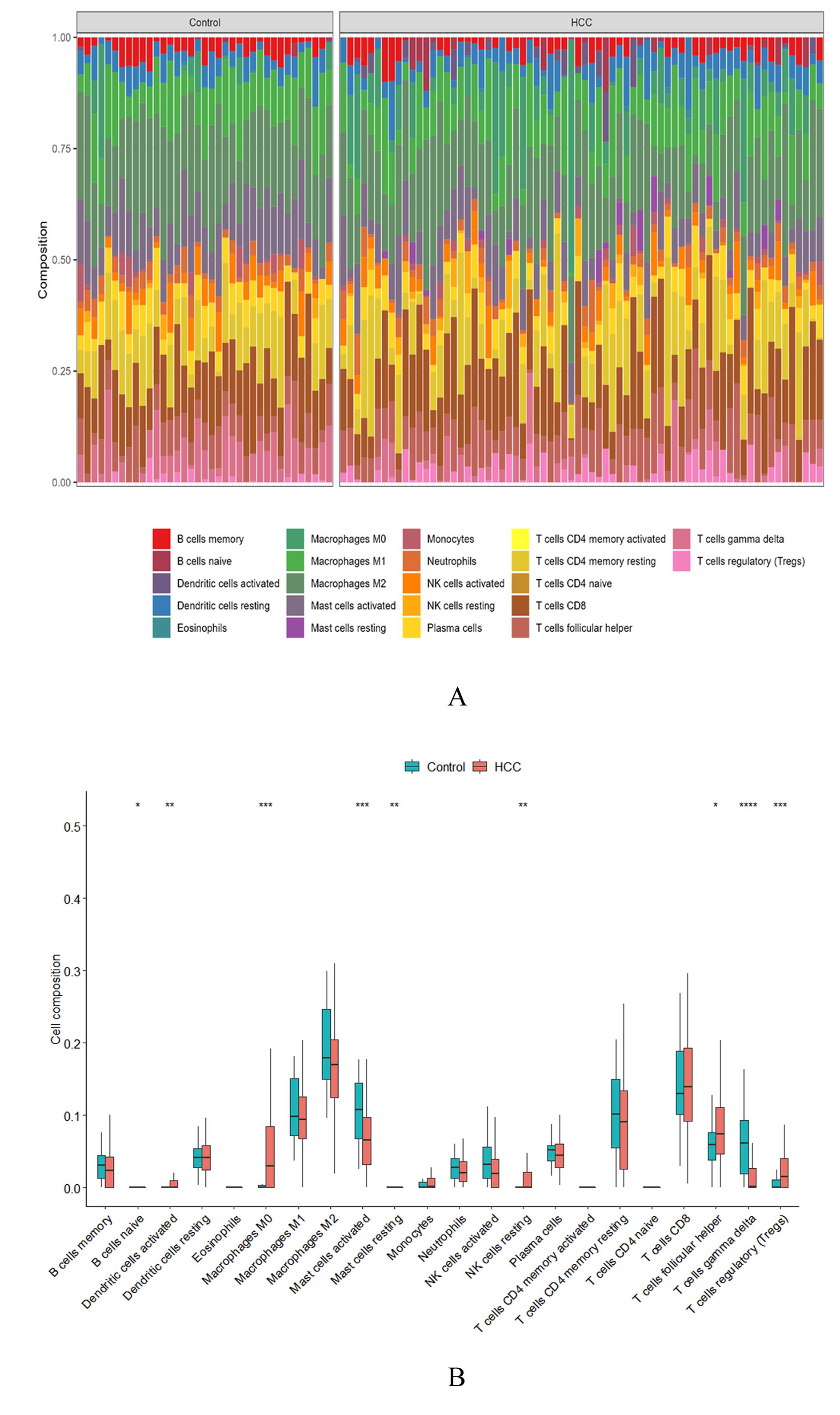
| 5 | DING W, ZHANG Z, YE N Y, et al. Identification of key genes in the HBV-related HCC immune microenvironment using integrated bioinformatics analysis[J]. J Oncol, 2022, 2022: 2797033. |
| 6 | WANG S M, OOI L L P J, HUI K M. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma[J]. Clin Cancer Res, 2007, 13(21): 6275-6283. |
| 7 | SZKLARCZYK D, GABLE A L, NASTOU K C,et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets[J]. Nucleic Acids Res, 2021, 49(D1): D605-D612. |
| 8 | CHIN C H, CHEN S H, WU H H, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome[J]. BMC Syst Biol, 2014, 8(): S11. |
| 9 | TANG Z F, LI C W, KANG B X, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[J]. Nucleic Acids Res, 2017, 45(W1): W98-W102. |
| 10 | LÁNCZKY A, GYÖRFFY B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation[J]. J Med Internet Res, 2021, 23(7): e27633. |
| 11 | ASPLUND A, EDQVIST P H D, SCHWENK J M, et al. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research[J]. Proteomics, 2012, 12(13): 2067-2077. |
| 12 | YANG J D, HAINAUT P, GORES G J, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604. |
| 13 | SCHWEITZER A, HORN J, MIKOLAJCZYK R T, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013[J]. Lancet, 2015, 386(10003): 1546-1555. |
| 14 | REVILL P A, TU T, NETTER H J, et al. The evolution and clinical impact of hepatitis B virus genome diversity[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(10): 618-634. |
| 15 | COLOTTA F, ALLAVENA P, SICA A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability[J]. Carcinogenesis, 2009, 30(7): 1073-1081. |
| 16 | HUANG X B, HE Y G, ZHENG L, et al. Identification of hepatitis B virus and liver cancer bridge molecules based on functional module network[J]. World J Gastroenterol, 2019, 25(33): 4921-4932. |
| 17 | PETTINELLI P, ARENDT B M, TETERINA A,et al.Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease[J]. PLoS One, 2018, 13(10): e0205747. |
| 18 | GAO J, WANG Z, WANG G J, et al. From hepatofibrosis to hepatocarcinogenesis: higher cytochrome P450 2E1 activity is a potential risk factor[J].Mol Carcinog, 2018, 57(10): 1371-1382. |
| 19 | OLIVEIRA P A, COLAÇO A, CHAVES R, et al. Chemical carcinogenesis[J]. An Acad Bras Cienc, 2007, 79(4): 593-616. |
| 20 | RAJAGOPALAN H, LENGAUER C. Aneuploidy and cancer[J]. Nature, 2004, 432(7015): 338-341. |
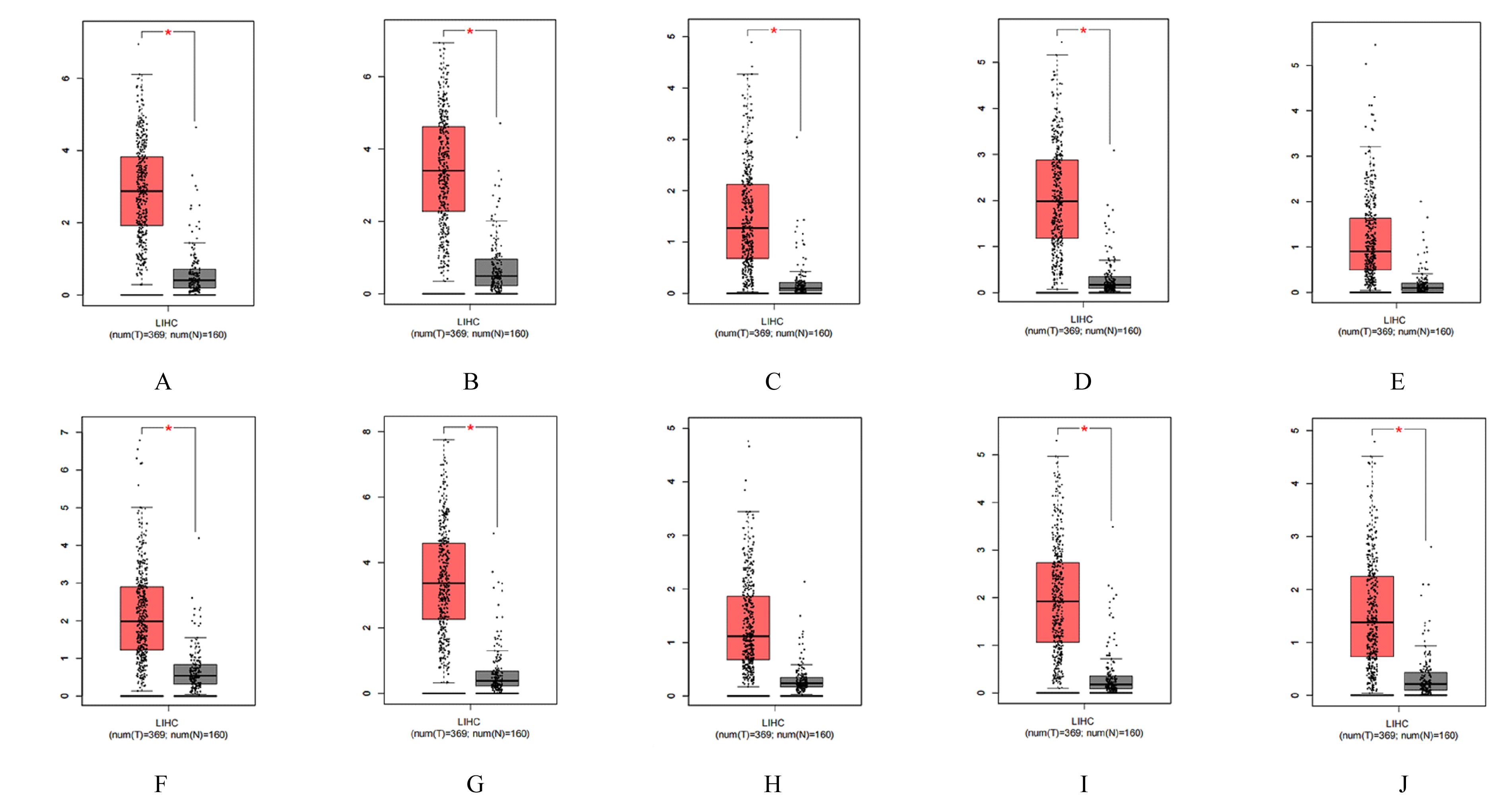
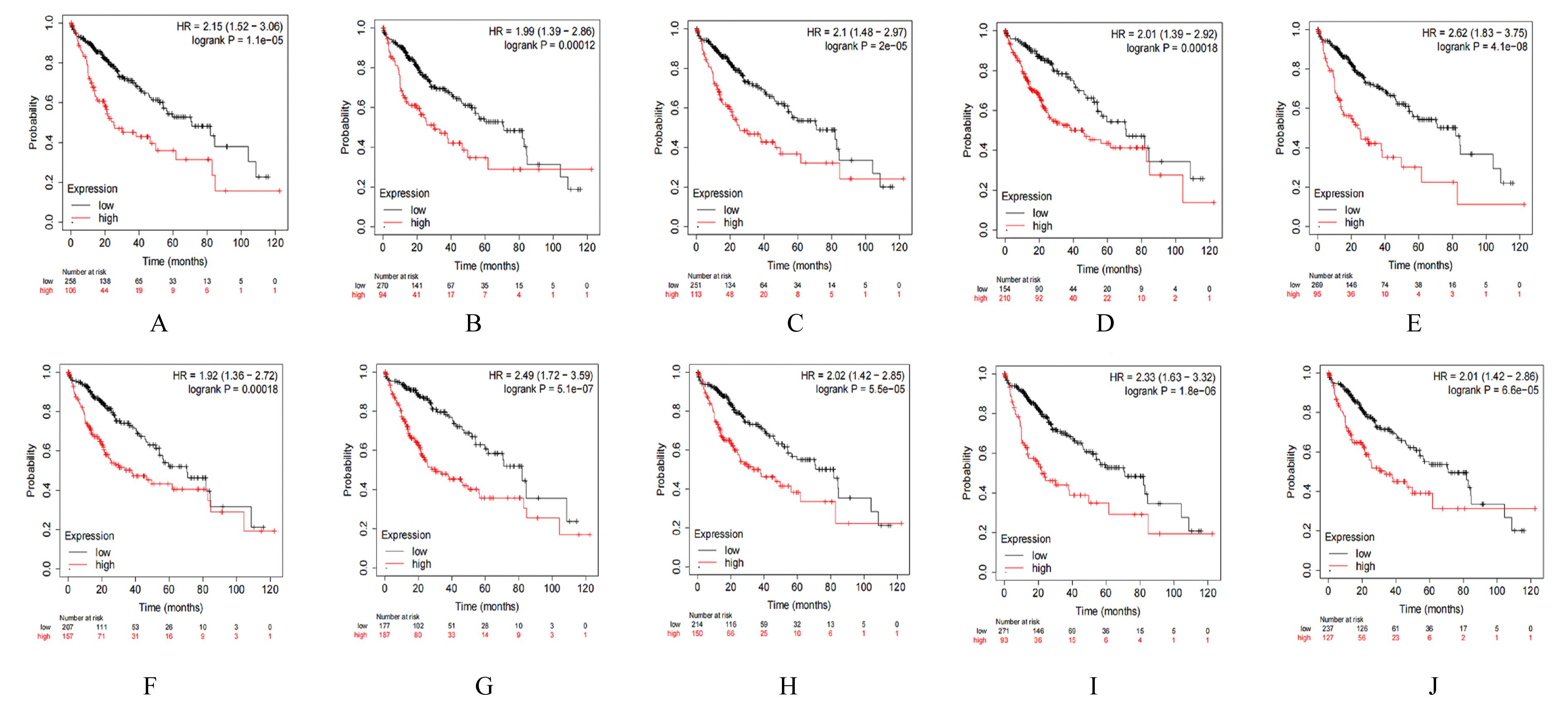
| 21 | ZHAO J, HAN S X, MA J L, et al. The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells[J]. Oncol Rep, 2013, 30(1): 253-259. |
| 22 | ITO Y, TAKEDA T, SAKON M, et al. Expression and prognostic role of cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma[J]. Oncology, 2000,59(1): 68-74. |
| 23 | KIM D H, PARK S E, KIM M, et al. A functional single nucleotide polymorphism at the promoter region of cyclin A2 is associated with increased risk of colon, liver, and lung cancers[J]. Cancer, 2011, 117(17): 4080-4091. |
| 24 | MA Q, LIU Y M, SHANG L, et al. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma[J]. Oncol Rep, 2017, 38(6): 3367-3375. |
| 25 | NG I O, NA J, LAI E C, et al. Ki-67 antigen expression in hepatocellular carcinoma using monoclonal antibody MIB1. A comparison with proliferating cell nuclear antigen[J]. Am J Clin Pathol, 1995, 104(3): 313-318. |
| 26 | KUO T C, CHANG P Y, HUANG S F, et al. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53[J]. Biochem Pharmacol, 2012, 83(6): 758-768. |
| 27 | HE B S, YIN J B, GONG S C, et al. Bioinformatics analysis of key genes and pathways for hepatocellular carcinoma transformed from cirrhosis[J]. Medicine, 2017, 96(25): e6938. |
| 28 | WU B, HU C Y, KONG L B. ASPM combined with KIF11 promotes the malignant progression of hepatocellular carcinoma via the Wnt/β-catenin signaling pathway[J]. Exp Ther Med, 2021, 22(4): 1154. |
| 29 | JEFFERY J, SINHA D, SRIHARI S, et al. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis[J]. Oncogene, 2016, 35(6): 683-690. |
| 30 | GASNEREAU I, BOISSAN M, MARGALL-DUCOS G, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis[J].Am J Pathol, 2012,180(1):131-140. |
| 31 | GUIDOTTI L G, CHISARI F V. Noncytolytic control of viral infections by the innate and adaptive immune response[J]. Annu Rev Immunol, 2001, 19: 65-91. |
| 32 | THIMME R, WIELAND S, STEIGER C, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection[J]. J Virol, 2003, 77(1): 68-76. |
| 33 | YANG P Y, LI Q J, FENG Y X, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma[J]. Cancer Cell, 2012, 22(3): 291-303. |
| 34 | JIN Z, SUN R, WEI H, et al. Accelerated liver fibrosis in hepatitis B virus transgenic mice: involvement of natural killer T cells[J].Hepatology,2011,53(1):219-229. |
| 35 | LI X, SU Y, HUA X, et al. Levels of hepatic Th17 cells and regulatory T cells upregulated by hepatic stellate cells in advanced HBV-related liver fibrosis[J]. J Transl Med,2017, 15(1):75. |
| 36 | KONG X, SUN R, CHEN Y, et al. γδT cells drive myeloid-derived suppressor cell-mediated CD8+ T cell exhaustion in hepatitis B virus-induced immunotolerance[J]. J Immunol,2014, 193(4):1645-1653. |
| [1] | Wenjun DENG,Liantao HU,Binnan ZHAO,Xinyu DONG,Xuebin LI,Jie LI,Xinyan YANG,Xiaoli GUO,Yue LI,Yikun QU,Weiqun WANG. Effect of CXC chemokine ligand 10 on proliferation and migration of hepatocellular carcinoma SMMC-7721 cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1227-1233. |
| [2] | Yong DONG,Lingyao XU,Jing HUA,Han LIANG,Dongya LIU,Junbo ZHAO,Zhenglu SUN,Cheng CHENG,Shutang WEI. Effect of macrophage exosomal lncRNA HULC on migration, invasion,and metastasis of hepatocellular carcinoma cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1217-1226. |
| [3] | Yuanyuan LIANG,Song ZHAO,Jing HU,Ni AN,Yanlu WEI,Rongjian SU. Effect of motesanib combined with EZH2 inhibitor GSK126 on proliferation and apoptosis of liver cancer Huh7 cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 896-904. |
| [4] | Jingjing LI, REZIWANGULI·Aisikaier,Wanyi XING,Yinggang ZOU. Primary dedifferentiated liposarcoma of unilateral ovary: A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 1040-1045. |
| [5] | Renyi YANG,Shuwang PENG,Yongheng WANG,Yuxuan DONG,Shanshan DUAN. Construction of ferroptosis prognostic risk model of thyroid cancer and bioinformatics analysis on its potential mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 402-413. |
| [6] | Xiaoyan WANG,Yihong HU,Yucheng HAN,Xianqiong ZOU. Bioinformatics analysis on mechanism of COMMD7 in occurrence and development of brain low-grade glioma [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 414-424. |
| [7] | Rui WANG,Ding ZHANG,Ruijian ZHUGE,Qian XUE,Li GUO. Bioinformatics analysis on mechanism of liver injury induced by hexavalent chromium [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 452-459. |
| [8] | Zhiyuan XIAO,Bing SONG,Xinyu MA,Lianhui JIN,Tong ZHENG,Fang CHAI. Bioinformatics analysis on expression of apoptosis related gene CD44 in thyroid carcinoma tissue and its relationship with tumor invasion and immune cell infiltration [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 473-481. |
| [9] | Yang BAI,Chenxi YANG,Xiaoqiang LIU,Yu LIU,Qian XING. Changes of follicular helper T lymphocytes in peripheral blood of patients with systemic lupus erythematosus during pregnancy and its significance [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 166-172. |
| [10] | Junyi JIN,Mu LI,Yaoyuan HU,Yihui WANG. Targeting relationship between miR-150-5p and NCAPG and its inhibitory effect on hepatocellular carcinoma Huh7 cells [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 122-130. |
| [11] | Wuyue TANG,Shuojie LI,Lijuan PANG. Bioinformatics analysis of splicing factor-alternative splicing regulatory network based on alternative splicing data in lung adenocarcinoma tissue [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 139-149. |
| [12] | Jilin FAN,Tingting ZHU,Xiaoling TIAN,Sijia LIU,Jing SU,Shiliang ZHANG. Bioinformatics analysis based on circRNA-miRNA-mRNA network construction and immune cell infiltration in atrial fibrillation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1535-1545. |
| [13] | Xiaoyan WANG,Qiuyue ZHANG,Yujie HU,Zhijing MO. Bioinformatics analysis based on molecular characteristics and mechanism of cystic fibrosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1290-1297. |
| [14] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
| [15] | Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765. |
|
||