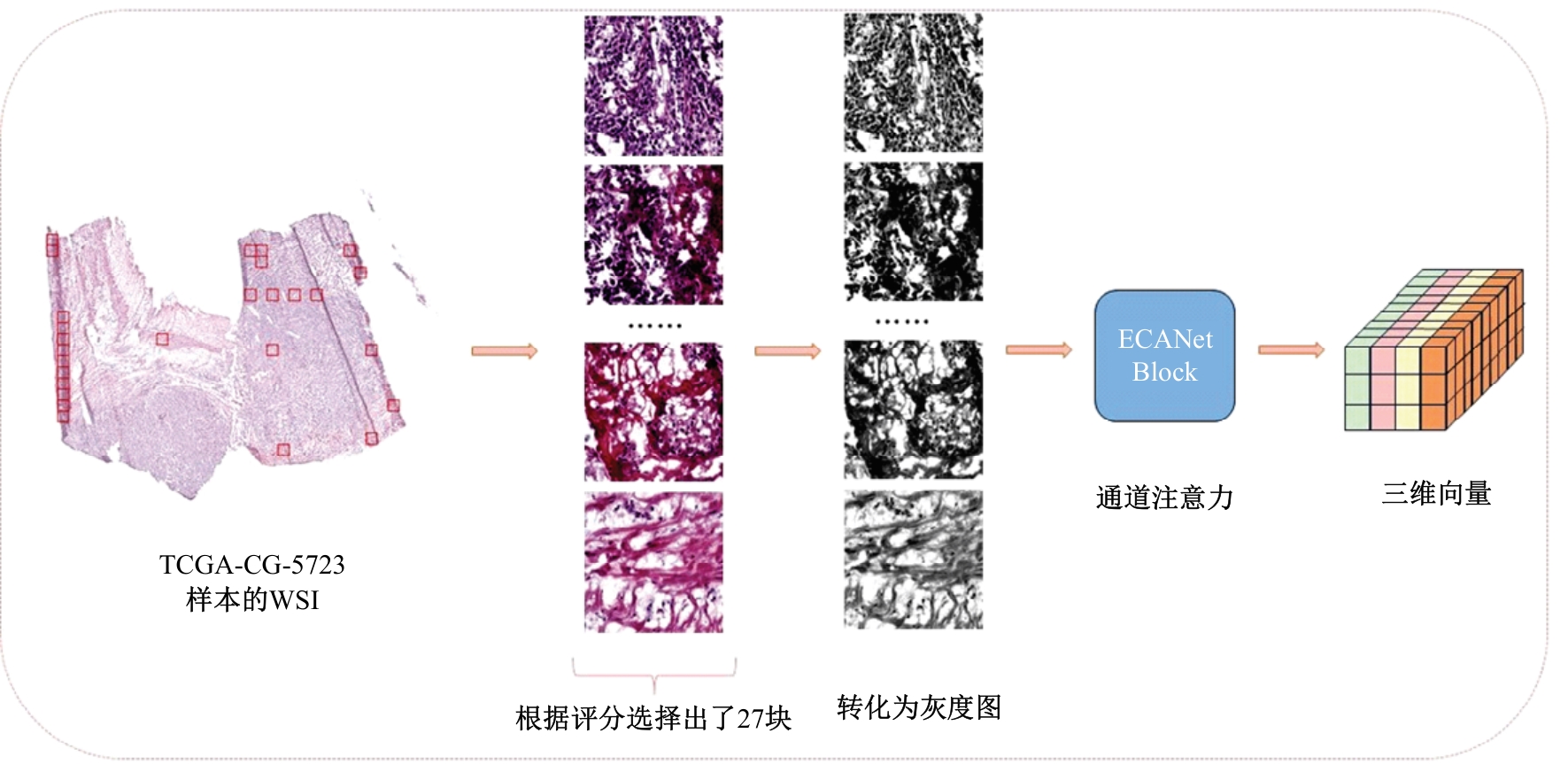
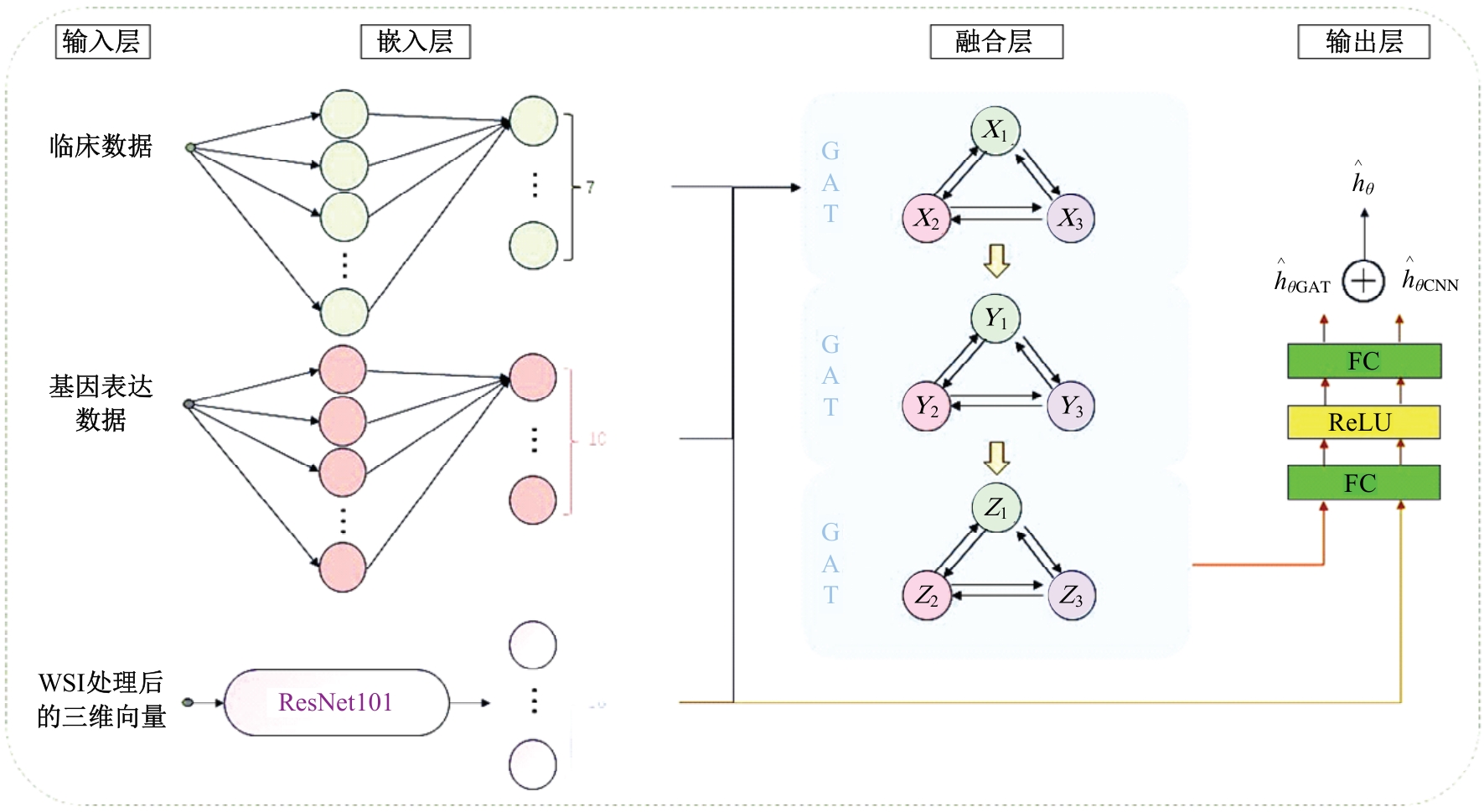
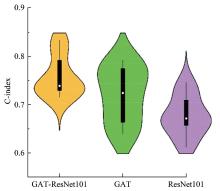
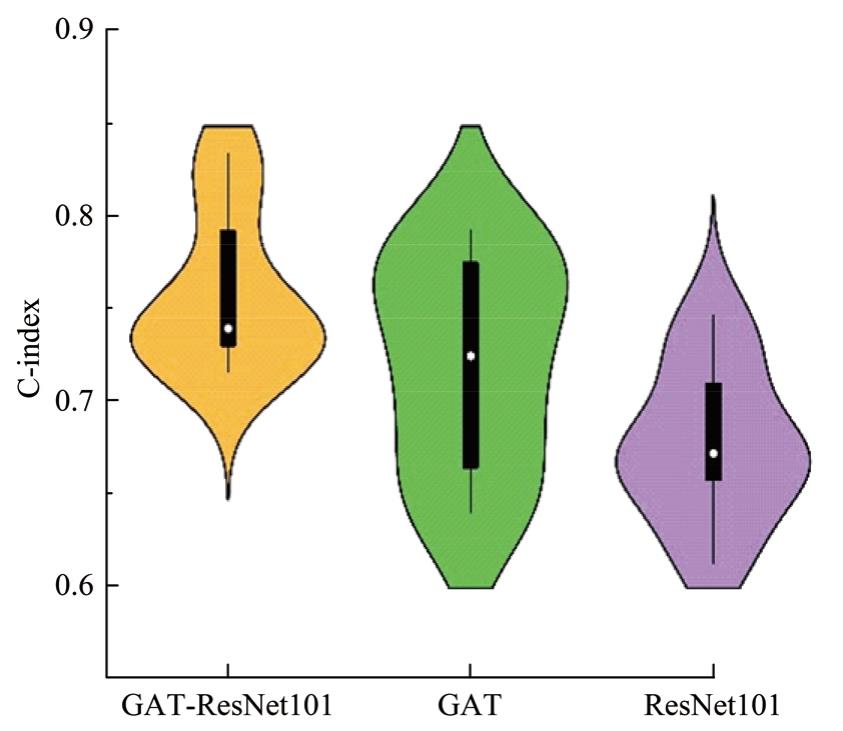
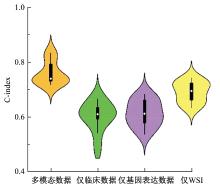
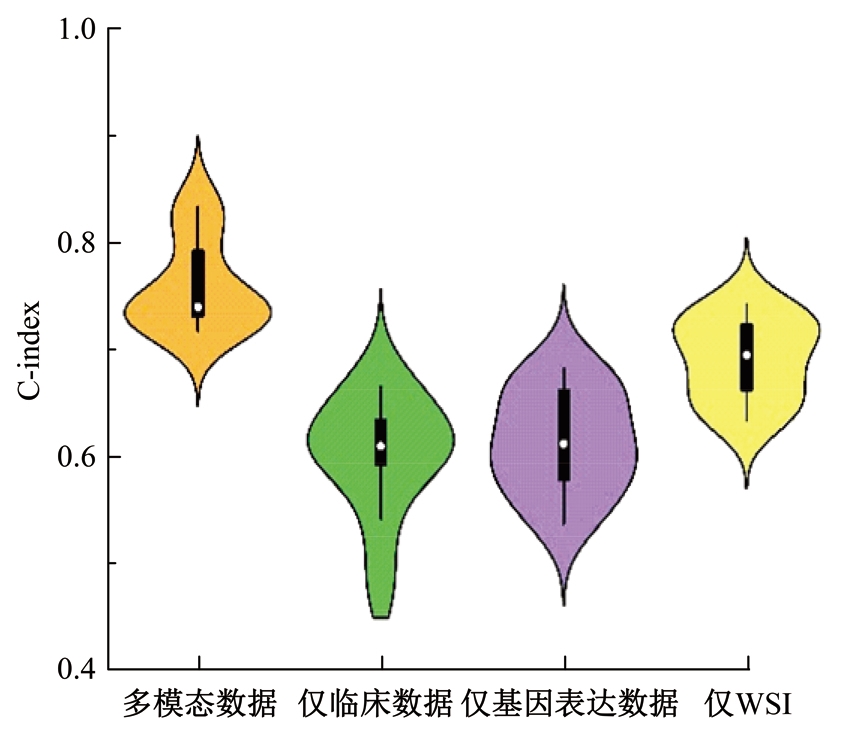
Journal of Jilin University(Engineering and Technology Edition) ›› 2025, Vol. 55 ›› Issue (8): 2693-2702.doi: 10.13229/j.cnki.jdxbgxb.20231262
Stomach cancer survival prediction model based on multimodal data fusion
Yuan-ning LIU1,2( ),Xing-zhe WANG1,2,Zi-yu HUANG3,Jia-chen ZHANG1(
),Xing-zhe WANG1,2,Zi-yu HUANG3,Jia-chen ZHANG1( ),Zhen LIU1,4
),Zhen LIU1,4
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education,Jilin University,Changchun 130012,China
3.School of Information Science and Technology,Beijing Forestry University,Beijing 100083,China
4.Graduate School of Engineering,Nagasaki Institute of Applied Science,Nagasaki 851-0193,Japan
CLC Number:
- TP391
| [1] | Huang S, Yang J, Fong S, et al. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges[J]. Cancer Letters, 2020, 471: 61-71. |
| [2] | Sung H, Ferlay J, Siegel R L, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2021, 71(3): 209-249. |
| [3] | Guan W L, He Y, Xu R H. Gastric cancer treatment: recent progress and future perspectives[J]. Journal of Hematology & Oncology, 2023, 16(1): 57-84. |
| [4] | 覃丽粒, 马小波, 赵天业, 等. MMP-9和TIMP-1表达在胃癌根治术后患者预后评估中的作用[J]. 吉林大学学报: 医学版, 2022, 48(1): 163-171. |
| Qin Li-li, Ma Xiao-bo, Zhao Tian-ye, et al. Effects of MMP-9 and TIMP-1 expressions on prognostic evaluation of gastric cancer patients after radical gastrectomy[J]. Journal of Jilin University (Medicine Edition), 2022, 48(1): 163-171. | |
| [5] | 崔海康, 张旭东, 李晓宁, 等. 细胞焦亡分型和APOD 预测胃癌患者预后作用的生物信息学分析[J]. 吉林大学学报: 医学版, 2023, 49(5): 1268-1279. |
| Cui Hai-kang, Zhang Xu-dong, Li Xiao-ning, et al. Bioinformatics analysis on predition effect of subtypes of cell pyroptosis and APOD on prognosis of gastric cancer patients[J]. Journal of Jilin University (Medicine Edition), 2023, 49(5): 1268-1279. | |
| [6] | Boorn H G, Engelhardt E G, Kleef J, et al. Prediction models for patients with esophageal or gastric cancer: a systematic review and meta-analysis[J]. Plos One, 2018, 13(2): No.e0192310. |
| [7] | Hosny A, Parmar C, Quackenbush J, et al. Artificial intelligence in radiology[J]. Springer Science and Business Media LLC, 2018, 18: 500-510. |
| [8] | Tran K A, Kondrashova O, Bradley A, et al. Deep learning in cancer diagnosis, prognosis and treatment selection[J]. Genome Medicine, 2021, 13(1): 152-168. |
| [9] | Krizhevsky A, Sutskever I, Hinton G E. ImageNet classification with deep convolutional neural networks[J]. Communications of the ACM, 2017, 60(6): 84-90. |
| [10] | Lecun Y, Bengio Y, Hinton G. Deep learning[J]. Nature, 2015, 521: 436-444. |
| [11] | Katzman J L, Shaham U, Cloninger A, et al. Deepsurv: personalized treatment recommender system using a cox proportional hazards deep neural network[J]. BMC Medical Research Methodology, 2018, 18(1): 24-35. |
| [12] | Choi S, Kim S. Artificial intelligence in the pathology of gastric cancer[J]. Journal of Gastric Cancer, 2023, 23(3): 410-427. |
| [13] | Deepa P, Gunavathi C. A systematic review on machine learning and deep learning techniques in cancer survival prediction[J]. Progress in Biophysics and Molecular Biology, 2022, 174: 62-71. |
| [14] | Lipkova J, Chen R J, Chen B, et al. Artificial intelligence for multimodal data integration in oncology[J]. Cancer Cell, 2022, 40(10): 1095-1110. |
| [15] | Boehm K M, Khosravi P, Vanguri R, et al. Harnessing multimodal data integration to advance precision oncology[J]. Nature Reviews Cancer, 2021, 22(2): 114-126. |
| [16] | Wei T, Yuan X, Gao R, et al. Survival prediction of stomach cancer using expression data and deep learning models with histopathological images[J]. Cancer Science, 2022, 114(2): 690-701. |
| [17] | Cox D R. Regression models and life-tables[J]. Journal of the Royal Statistical Society, Series B (Statistical Methodology), 1972, 34(2): 187-202. |
| [18] | Cox D R, Oakes D. Analysis of survival data[J]. Biometrics, 1985, 41(2): 593-594. |
| [19] | Tarkhan A, Simon N, Bengtsson T, et al. Survival prediction using deep learning[C]∥Proceedings of AAAI Spring Symposium on Survival Prediction–Algorithms, Challenges, and Applications, Virtual, USA, 2021: 1-8. |
| [20] | Li H, Lin D, Yu Z, et al. A nomogram model based on the number of examined lymph nodes–related signature to predict prognosis and guide clinical therapy in gastric cancer[J]. Frontiers in Immunology, 2022, 13: No.947802. |
| [21] | Wu J, Wang X, Wang N, et al. Identification of novel antioxidant gene signature to predict the prognosis of patients with gastric cancer[J]. World Journal of Surgical Oncology, 2021, 19(1): No.1901219. |
| [22] | Dai W, Xiao Y, Tang W, et al. Identification of an emt-related gene signature for predicting overall survival in gastric cancer[J]. Frontiers in Genetics, 2021, 12: No. 661306. |
| [23] | Wang M, Jing J, Li H, et al. The expression characteristics and prognostic roles of autophagy-related genes in gastric cancer[J]. PeerJ, 2021, 9: No.e10814. |
| [24] | Liu M, Li J, Huang Z, et al. Gastric cancer risk-scoring system based on analysis of a competing endogenous RNA network[J]. Translational Cancer Research, 2020, 9(6): 3889-3902. |
| [25] | Kourou K, Exarchos T P, Exarchos K P, et al. Machine learning applications in cancer prognosis and prediction[J]. Computational and Structural Biotechnology Journal, 2015, 13: 8-17. |
| [26] | Shivaswamy P K, Chu W, Jansche M. A support vector approach to censored targets[C]∥ Proceedings of Seventh IEEE International Conference on Data Mining, Omaha, USA, 2007: 655-660. |
| [27] | Ishwaran H, Kogalur U B, Blackstone E H, et al. Random survival forests[J]. Annals of Applied Statistics, 2008, 2: No.AOAS169. |
| [28] | Chen T, Guestrin C. XGBoost: a scalable tree boosting system[C]∥Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, USA, 2016: 785-794. |
| [29] | Liu P, Fu B, Yang S X, et al. Optimizing survival analysis of xgboost for ties to predict disease progression of breast cancer[J]. Institute of Electrical and Electronics Engineers, 2021, 68: 148-160. |
| [30] | Ma B, Yan G, Chai B, et al. XGBLC: an improved survival prediction model based on XGBoost[J]. Bioinformatics, 2021, 38(2): 410-418. |
| [31] | Li G, Huo D, Guo N, et al. Integrating multiple machine learning algorithms for prognostic prediction of gastric cancer based on immune-related lncRNAs[J]. Frontiers in Genetics, 2023, 14: No.1106724. |
| [32] | Chai H, Zhou X, Zhang Z, et al. Integrating multi-omics data through deep learning for accurate cancer prognosis prediction[J]. Computers in Biology and Medicine, 2021, 134: No.104481. |
| [33] | Wang Y, Zhang Z, Chai H, et al. Multi-omics cancer prognosis analysis based on graph convolution network[C]∥Proceedings of IEEE International Conference on Bioinformatics and Biomedicine, Houston, USA, 2021: 1564-1568. |
| [34] | Zhang Y, Xiong S, Wang Z, et al. Local augmented graph neural network for multi-omics cancer prognosis prediction and analysis[J]. Methods, 2023, 213: 1-9. |
| [35] | Avelar P H C, Tavares A R, Silveira T L T, et al. Superpixel image classification with graph attention networks[C]∥Proceedings of 33rd SIBGRAPI Conference on Graphics, Patterns and Images, Porto de Galinhas, Brazil, 2020: 203-209. |
| [36] | Velickovic P, Cucurull G, Casanova A, et al. Graph Attention Networks[EB/OL]. [2023-11-04]. . |
| [37] | Fu X, Patrick E, Yang J Y H, et al. Deep multimodal graph-based network for survival prediction from highly multiplexed images and patient variables[J]. Computers in Biology and Medicine, 2023, 154: No.106576. |
| [38] | Duan M, Wang Y, Zhao D, et al. Orchestrating information across tissues via a novel multitask GAT framework to improve quantitative gene regulation relation modeling for survival analysis[J]. Briefings in Bioinformatics, 2023, 24(4): 1-10. |
| [39] | Ye L, Zhang Y, Yang X, et al. An ovarian cancer susceptible gene prediction method based on deep learning methods[J]. Frontiers in Cell and Developmental Biology, 2021, 9: No.730475. |
| [40] | Adeoye J, Hui L, Koohi M M, et al. Comparison of time-to-event machine learning models in predicting oral cavity cancer prognosis[J]. International Journal of Medical Informatics, 2022, 157: No.104635. |
| [41] | Lerademacher J, Wang X. Time-to-event data: an overview and analysis considerations[J]. Journal of Thoracic Oncology, 2021, 16(7): 1067-1074. |
| [42] | Amin M B, Greene F L, Edge S B, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging[J]. CA-A Cancer Journal for Clinicians, 2017, 67(2): 93-99. |
| [43] | Marcolini A, Bussola N, Arbitrio E, et al. Histolab: a python library for reproducible digital pathology preprocessing with automated testing[J]. Software X, 2020, 20: No.101237. |
| [44] | He K, Zhang X, Ren S, et al. Deep residual learning for image recognition[C]∥Proceedings of 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 770-778. |
| [45] | Brentnall A R, Cuzick J. Use of the concordance index for predictors of censored survival data[J]. Statistical Methods in Medical Research, 2018, 27(8): 2359-2373. |
| [46] | Kaplan E L, Meier P. Nonparametric estimation from incomplete observations[J]. Journal of the American Statistical Association, 1958, 53(282): 457-481. |
| [47] | Pan Q K, Xu X X, Qi C, et al. Feature fusion: graph attention network and cnn combing for hyperspectral image classification[C]∥Proceedings of the 5th International Conference on Control and Computer Vision, New York, USA, 2022: 171-178. |
| [48] | Xie Y, Niu G, Da Q, et al. Survival prediction for gastric cancer via multimodal learning of whole slide images and gene expression[C]∥Proceedings of 2022 IEEE International Conference on Bioinformatics and Biomedicine, Las Vegas, USA, 2022: 1311-1316. |
| [49] | 张德洪, 郑明珠, 李家秋, 等. 基于MSR1 mRNA 和蛋白在泛癌组织中表达的生物信息学分析及其意义[J]. 吉林大学学报: 医学版, 2023, 49(2): 425-439. |
| Zhang De-hong, Zheng Ming-zhu, Li Jia-qiu, et al. Bioinformatics analysis based on expressions of MSR1 mRNA and protein in pan-cancer tissue and its significance[J]. Journal of Jilin University (Medicine Edition), 2023, 49(2): 425-439. | |
| [50] | Selvaraju R R, Cogswell M, Das A, et al. Grad-CAM:visual explanations from deep networks via gradient-based localization[C]∥2017 IEEE International Conference on Computer Vision, Venice, Italy, 2017: 618-626. |
| [51] | Krzyziński M, Spytek M, Baniecki H, et al. Survshap(t): time-dependent explanations of machine learning survival models[J]. Knowledge-Based Systems, 2023, 262: 110234. |
| [1] | Jing-shu YUAN,Wu LI,Xing-yu ZHAO,Man YUAN. Semantic matching model based on BERTGAT-Contrastive [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(7): 2383-2392. |
| [2] | Hui-zhi XU,Dong-sheng HAO,Xiao-ting XU,Shi-sen JIANG. Expressway small object detection algorithm based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(6): 2003-2014. |
| [3] | Ru-bo ZHANG,Shi-qi CHANG,Tian-yi ZHANG. Review on image information hiding methods based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1497-1515. |
| [4] | Jian LI,Huan LIU,Yan-qiu LI,Hai-rui WANG,Lu GUAN,Chang-yi LIAO. Image recognition research on optimizing ResNet-18 model based on THGS algorithm [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1629-1637. |
| [5] | Bin WEN,Yi-fu DING,Chao YANG,Yan-jun SHEN,Hui LI. Self-selected architecture network for traffic sign classification [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(5): 1705-1713. |
| [6] | Zhen-jiang LI,Li WAN,Shi-rui ZHOU,Chu-qing TAO,Wei WEI. Dynamic estimation of operational risk of tunnel traffic flow based on spatial-temporal Transformer network [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(4): 1336-1345. |
| [7] | Meng-xue ZHAO,Xiang-jiu CHE,Huan XU,Quan-le LIU. A method for generating proposals of medical image based on prior knowledge optimization [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(2): 722-730. |
| [8] | Hui-zhi XU,Shi-sen JIANG,Xiu-qing WANG,Shuang CHEN. Vehicle target detection and ranging in vehicle image based on deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(1): 185-197. |
| [9] | Yuan-ning LIU,Zi-nan ZANG,Hao ZHANG,Zhen LIU. Deep learning-based method for ribonucleic acid secondary structure prediction [J]. Journal of Jilin University(Engineering and Technology Edition), 2025, 55(1): 297-306. |
| [10] | Lei ZHANG,Jing JIAO,Bo-xin LI,Yan-jie ZHOU. Large capacity semi structured data extraction algorithm combining machine learning and deep learning [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(9): 2631-2637. |
| [11] | Lu Li,Jun-qi Song,Ming Zhu,He-qun Tan,Yu-fan Zhou,Chao-qi Sun,Cheng-yu Zhou. Object extraction of yellow catfish based on RGHS image enhancement and improved YOLOv5 network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(9): 2638-2645. |
| [12] | Xin-gang GUO,Ying-chen HE,Chao CHENG. Noise-resistant multistep image super resolution network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(7): 2063-2071. |
| [13] | Bai-you QIAO,Tong WU,Lu YANG,You-wen JIANG. A text sentiment analysis method based on BiGRU and capsule network [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(7): 2026-2037. |
| [14] | Li-ping ZHANG,Bin-yu LIU,Song LI,Zhong-xiao HAO. Trajectory k nearest neighbor query method based on sparse multi-head attention [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1756-1766. |
| [15] | Ming-hui SUN,Hao XUE,Yu-bo JIN,Wei-dong QU,Gui-he QIN. Video saliency prediction with collective spatio-temporal attention [J]. Journal of Jilin University(Engineering and Technology Edition), 2024, 54(6): 1767-1776. |
|
||