| 1 |
BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J].CA Cancer J Clin,2018,68(6):394-424.
|
| 2 |
PETRICK J L, FLORIO A A, ZNAOR A, et al. International trends in hepatocellular carcinoma incidence, 1978-2012[J]. Int J Cancer, 2020, 147(2): 317-330.
|
| 3 |
RINALDI L, GUARINO M, PERRELLA A, et al. Role of liver stiffness measurement in predicting HCC occurrence in direct-acting antivirals setting: a real-life experience[J]. Dig Dis Sci, 2019, 64(10): 3013-3019.
|
| 4 |
GRANDHI M S, KIM A K, RONNEKLEIV-KELLY S M, et al. Hepatocellular carcinoma: from diagnosis to treatment[J]. Surg Oncol, 2016, 25(2): 74-85.
|
| 5 |
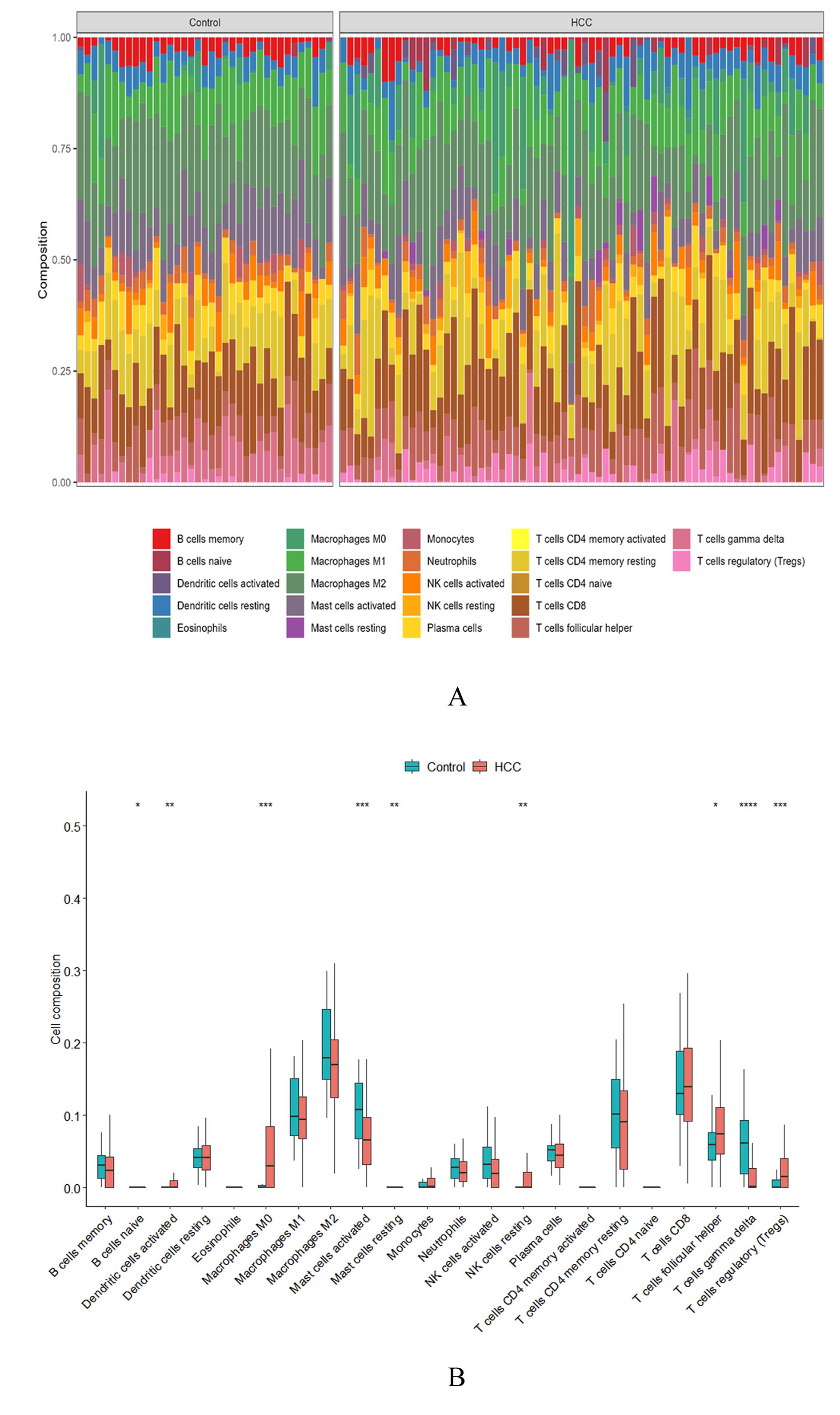
DING W, ZHANG Z, YE N Y, et al. Identification of key genes in the HBV-related HCC immune microenvironment using integrated bioinformatics analysis[J]. J Oncol, 2022, 2022: 2797033.
|
| 6 |
WANG S M, OOI L L P J, HUI K M. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma[J]. Clin Cancer Res, 2007, 13(21): 6275-6283.
|
| 7 |
SZKLARCZYK D, GABLE A L, NASTOU K C,et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets[J]. Nucleic Acids Res, 2021, 49(D1): D605-D612.
|
| 8 |
CHIN C H, CHEN S H, WU H H, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome[J]. BMC Syst Biol, 2014, 8(): S11.
|
| 9 |
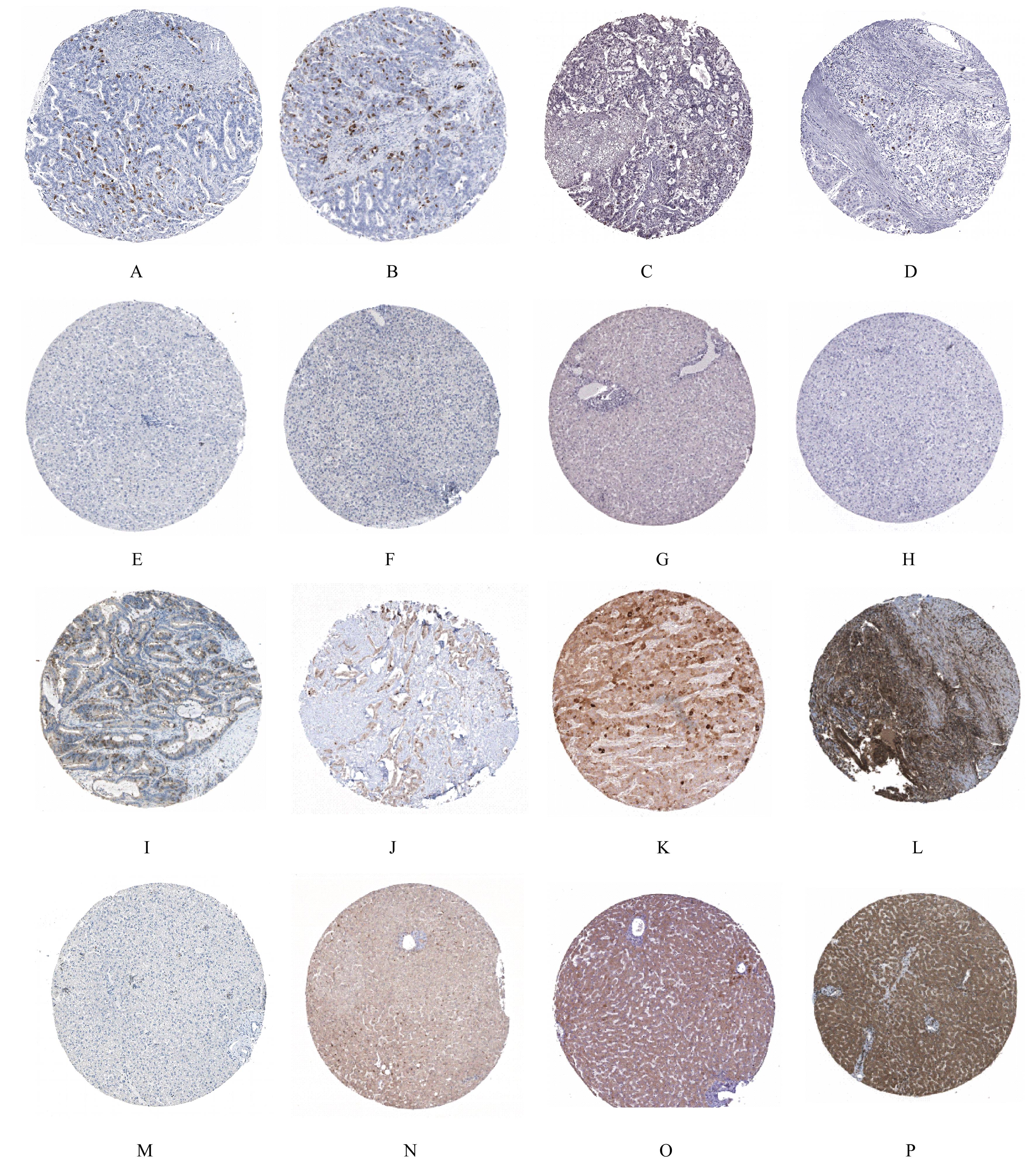
TANG Z F, LI C W, KANG B X, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[J]. Nucleic Acids Res, 2017, 45(W1): W98-W102.
|
| 10 |
LÁNCZKY A, GYÖRFFY B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation[J]. J Med Internet Res, 2021, 23(7): e27633.
|
| 11 |
ASPLUND A, EDQVIST P H D, SCHWENK J M, et al. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research[J]. Proteomics, 2012, 12(13): 2067-2077.
|
| 12 |
YANG J D, HAINAUT P, GORES G J, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604.
|
| 13 |
SCHWEITZER A, HORN J, MIKOLAJCZYK R T, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013[J]. Lancet, 2015, 386(10003): 1546-1555.
|
| 14 |
REVILL P A, TU T, NETTER H J, et al. The evolution and clinical impact of hepatitis B virus genome diversity[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(10): 618-634.
|
| 15 |
COLOTTA F, ALLAVENA P, SICA A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability[J]. Carcinogenesis, 2009, 30(7): 1073-1081.
|
| 16 |
HUANG X B, HE Y G, ZHENG L, et al. Identification of hepatitis B virus and liver cancer bridge molecules based on functional module network[J]. World J Gastroenterol, 2019, 25(33): 4921-4932.
|
| 17 |
PETTINELLI P, ARENDT B M, TETERINA A,et al.Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease[J]. PLoS One, 2018, 13(10): e0205747.
|
| 18 |
GAO J, WANG Z, WANG G J, et al. From hepatofibrosis to hepatocarcinogenesis: higher cytochrome P450 2E1 activity is a potential risk factor[J].Mol Carcinog, 2018, 57(10): 1371-1382.
|
| 19 |
OLIVEIRA P A, COLAÇO A, CHAVES R, et al. Chemical carcinogenesis[J]. An Acad Bras Cienc, 2007, 79(4): 593-616.
|
| 20 |
RAJAGOPALAN H, LENGAUER C. Aneuploidy and cancer[J]. Nature, 2004, 432(7015): 338-341.
|
| 21 |
ZHAO J, HAN S X, MA J L, et al. The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells[J]. Oncol Rep, 2013, 30(1): 253-259.
|
| 22 |
ITO Y, TAKEDA T, SAKON M, et al. Expression and prognostic role of cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma[J]. Oncology, 2000,59(1): 68-74.
|
| 23 |
KIM D H, PARK S E, KIM M, et al. A functional single nucleotide polymorphism at the promoter region of cyclin A2 is associated with increased risk of colon, liver, and lung cancers[J]. Cancer, 2011, 117(17): 4080-4091.
|
| 24 |
MA Q, LIU Y M, SHANG L, et al. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma[J]. Oncol Rep, 2017, 38(6): 3367-3375.
|
| 25 |
NG I O, NA J, LAI E C, et al. Ki-67 antigen expression in hepatocellular carcinoma using monoclonal antibody MIB1. A comparison with proliferating cell nuclear antigen[J]. Am J Clin Pathol, 1995, 104(3): 313-318.
|
| 26 |
KUO T C, CHANG P Y, HUANG S F, et al. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53[J]. Biochem Pharmacol, 2012, 83(6): 758-768.
|
| 27 |
HE B S, YIN J B, GONG S C, et al. Bioinformatics analysis of key genes and pathways for hepatocellular carcinoma transformed from cirrhosis[J]. Medicine, 2017, 96(25): e6938.
|
| 28 |
WU B, HU C Y, KONG L B. ASPM combined with KIF11 promotes the malignant progression of hepatocellular carcinoma via the Wnt/β-catenin signaling pathway[J]. Exp Ther Med, 2021, 22(4): 1154.
|
| 29 |
JEFFERY J, SINHA D, SRIHARI S, et al. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis[J]. Oncogene, 2016, 35(6): 683-690.
|
| 30 |
GASNEREAU I, BOISSAN M, MARGALL-DUCOS G, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis[J].Am J Pathol, 2012,180(1):131-140.
|
| 31 |
GUIDOTTI L G, CHISARI F V. Noncytolytic control of viral infections by the innate and adaptive immune response[J]. Annu Rev Immunol, 2001, 19: 65-91.
|
| 32 |
THIMME R, WIELAND S, STEIGER C, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection[J]. J Virol, 2003, 77(1): 68-76.
|
| 33 |
YANG P Y, LI Q J, FENG Y X, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma[J]. Cancer Cell, 2012, 22(3): 291-303.
|
| 34 |
JIN Z, SUN R, WEI H, et al. Accelerated liver fibrosis in hepatitis B virus transgenic mice: involvement of natural killer T cells[J].Hepatology,2011,53(1):219-229.
|
| 35 |
LI X, SU Y, HUA X, et al. Levels of hepatic Th17 cells and regulatory T cells upregulated by hepatic stellate cells in advanced HBV-related liver fibrosis[J]. J Transl Med,2017, 15(1):75.
|
| 36 |
KONG X, SUN R, CHEN Y, et al. γδT cells drive myeloid-derived suppressor cell-mediated CD8+ T cell exhaustion in hepatitis B virus-induced immunotolerance[J]. J Immunol,2014, 193(4):1645-1653.
|
 ),王素珍(
),王素珍( )
)
 ),Suzhen WANG(
),Suzhen WANG( )
)