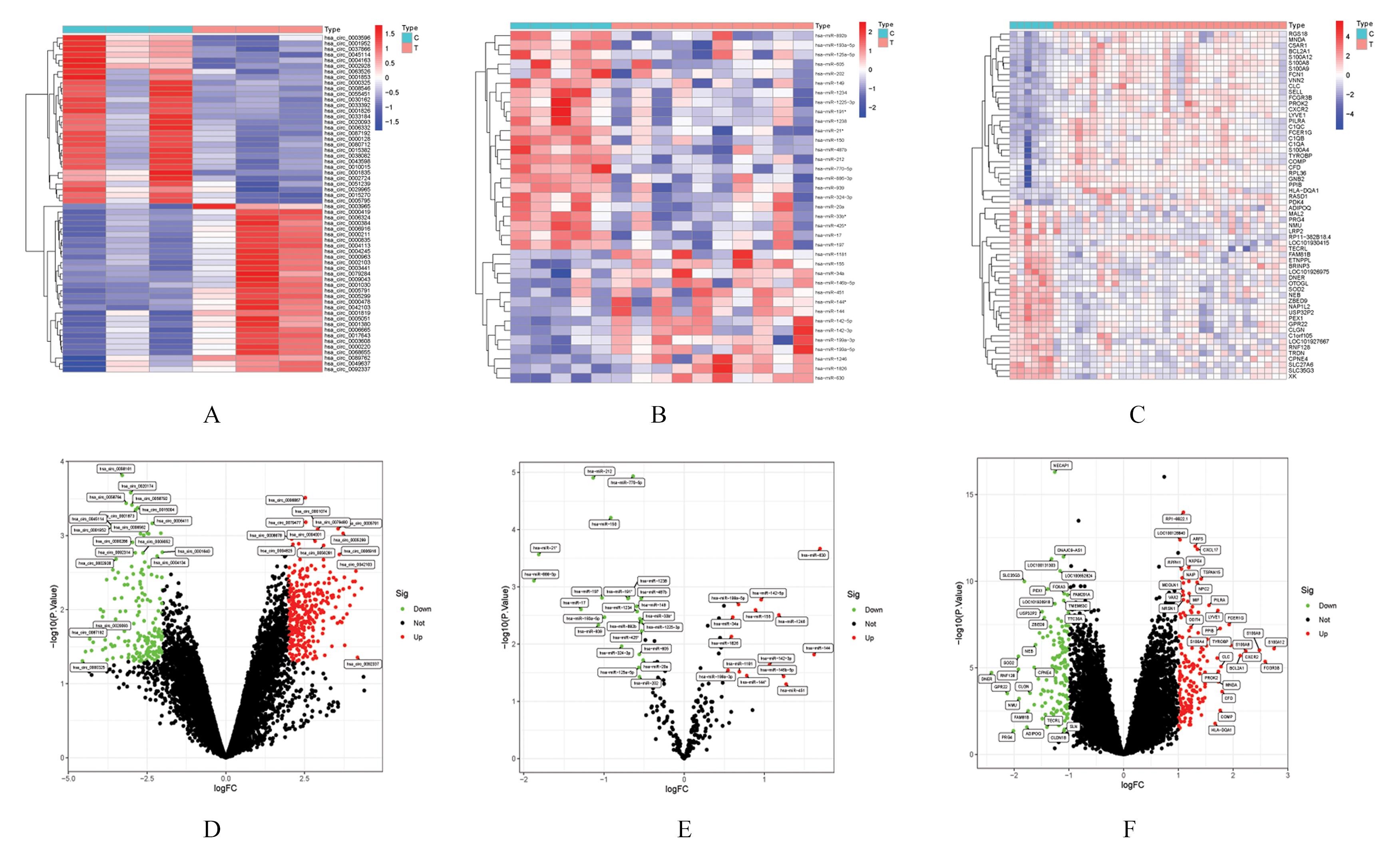
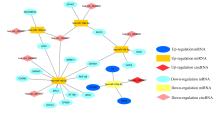
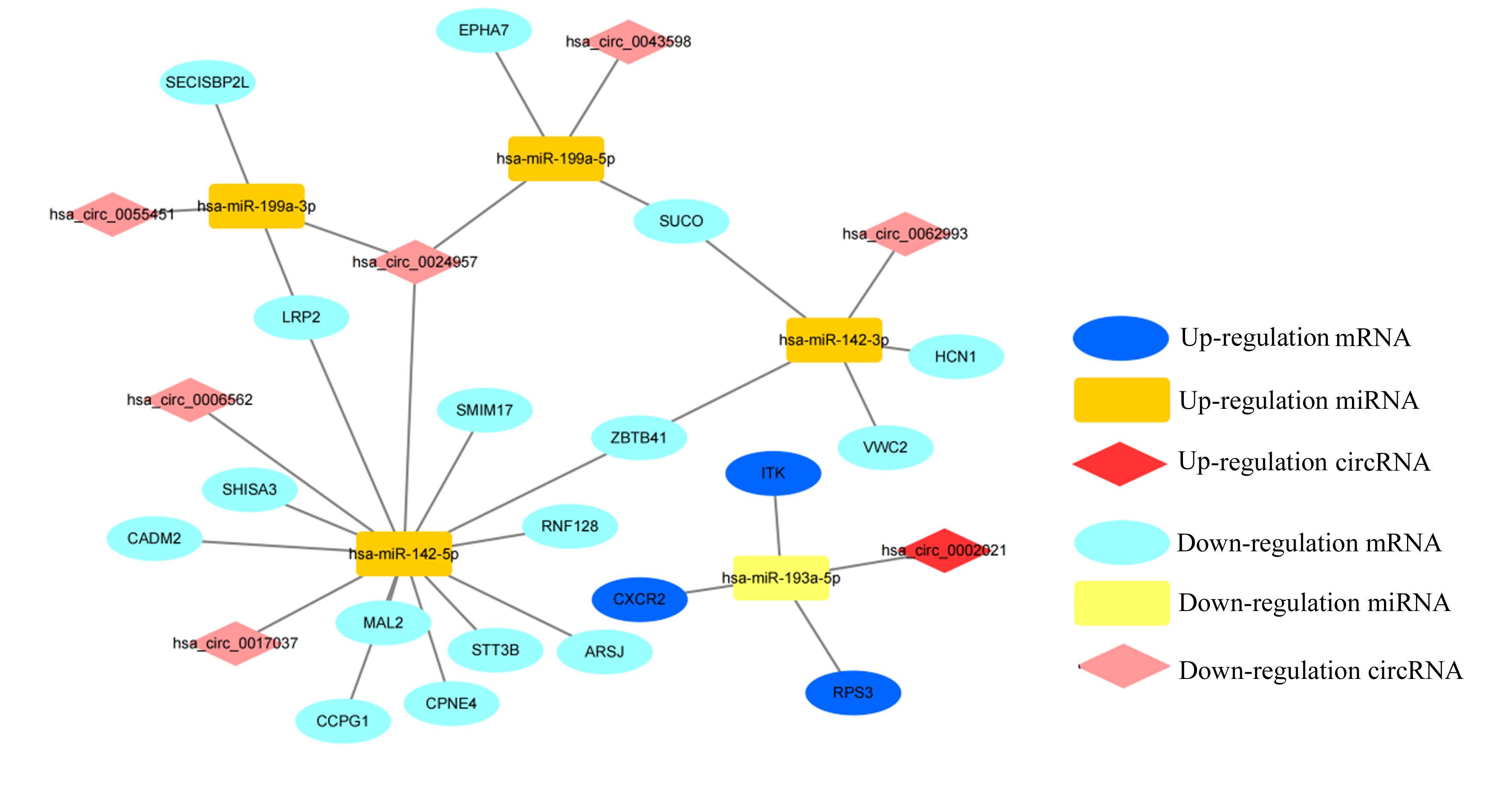
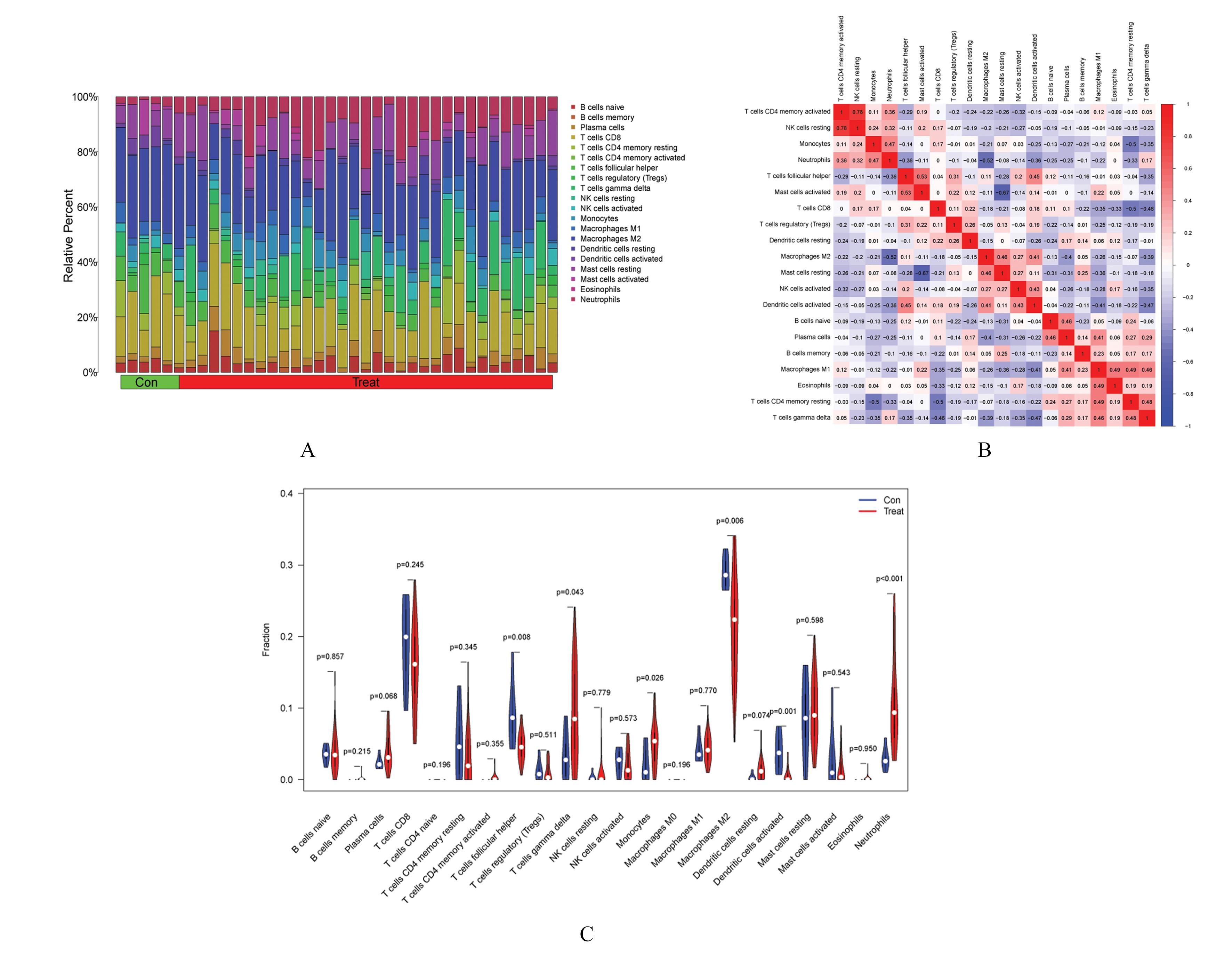
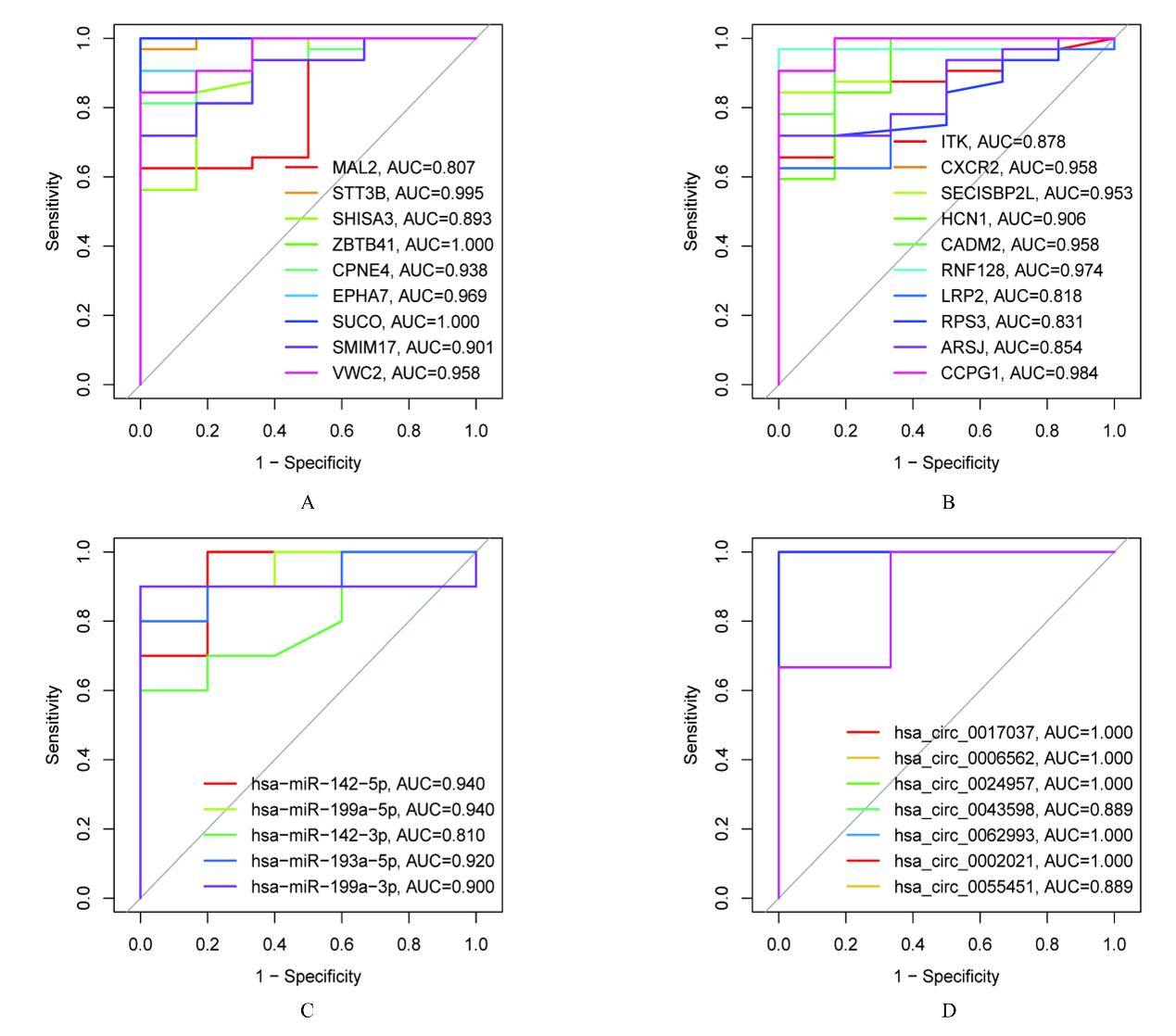
| 1 |
XIAO Y, WANG X Y, YANG J, et al. Huatan Dingji Decoction intervening in atrial fibrillation: protocol for a randomized double-blind single-simulated placebo-controlled clinical trial[J]. Trials, 2021, 22(1): 693.
|
| 2 |
LIU Y Z, SHI Q M, MA Y X, et al. The role of immune cells in atrial fibrillation[J]. J Mol Cell Cardiol, 2018, 123: 198-208.
|
| 3 |
CAO F, LI Z, DING W M,et al.Angiotensin Ⅱ-treated cardiac myocytes regulate M1 macrophage polarization via transferring exosomal PVT1[J]. J Immunol Res, 2021, 2021: 1994328.
|
| 4 |
MIYOSAWA K, IWATA H, MINAMI-TAKANO A, et al. Enhanced monocyte migratory activity in the pathogenesis of structural remodeling in atrial fibrillation[J]. PLoS One, 2020, 15(10): e0240540.
|
| 5 |
SZABO L, SALZMAN J. Detecting circular RNAs: bioinformatic and experimental challenges[J]. Nat Rev Genet, 2016, 17(11): 679-692.
|
| 6 |
BARRETT S P, SALZMAN J. Circular RNAs: analysis, expression and potential functions[J]. Development, 2016, 143(11): 1838-1847.
|
| 7 |
ZHANG X H, LU J Y, ZHANG Q H, et al. CircRNA RSF1 regulated ox-LDL induced vascular endothelial cells proliferation, apoptosis and inflammation through modulating miR-135b-5p/HDAC1 axis in atherosclerosis[J]. Biol Res, 2021, 54(1): 11.
|
| 8 |
TIAN T T, LI F, CHEN R H, et al. Therapeutic potential of exosomes derived from circRNA_0002113 lacking mesenchymal stem cells in myocardial infarction[J]. Front Cell Dev Biol, 2022, 9: 779524.
|
| 9 |
WANG K, LONG B, LIU F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223[J]. Eur Heart J, 2016, 37(33): 2602-2611.
|
| 10 |
SU Q, LV X W. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis[J]. Genomics, 2020, 112(2): 1680-1685.
|
| 11 |
WU N, LI C Y, XU B, et al. Circular RNA mmu_circ_0005019 inhibits fibrosis of cardiac fibroblasts and reverses electrical remodeling of cardiomyocytes[J]. BMC Cardiovasc Disord, 2021, 21(1): 308.
|
| 12 |
RITCHIE M E, PHIPSON B, WU D, et al. Limma Powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Res, 2015, 43(7): e47.
|
| 13 |
GHOSAL S, DAS S, SEN R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits[J]. Front Genet, 2013, 4: 283.
|
| 14 |
LI J H, LIU S, ZHOU H, et al. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data[J]. Nucl Acids Res, 2014, 42(D1): D92-D97.
|
| 15 |
LIU M, WANG Q, SHEN J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature[J]. RNA Biol, 2019, 16(7): 899-905.
|
| 16 |
AGARWAL V, BELL G W, NAM J W, et al. Predicting effective microRNA target sites in mammalian mRNAs[J]. eLife, 2015, 4: e05005.
|
| 17 |
WONG N, WANG X W.miRDB: an online resource for microRNA target prediction and functional annotations[J].Nucleic Acids Res,2015,43(Database issue): D146-D152.
|
| 18 |
HUANG D W, SHERMAN B T, LEMPICKI R A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009, 4(1): 44-57.
|
| 19 |
NEWMAN A M, LIU C L, GREEN M R, et al. Robust enumeration of cell subsets from tissue expression profiles[J].Nat Methods,2015,12(5): 453-457.
|
| 20 |
KRISTENSEN L S, ANDERSEN M S, STAGSTED L V W, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-691.
|
| 21 |
ZHANG Y L, TENG F, HAN X, et al. Selective blocking of CXCR2 prevents and reverses atrial fibrillation in spontaneously hypertensive rats[J]. J Cell Mol Med, 2020, 24(19): 11272-11282.
|
| 22 |
GROSSI I, SALVI A, ABENI E, et al. Biological function of microRNA193a-3p in health and disease[J]. Int J Genomics, 2017, 2017: 5913195.
|
| 23 |
HELLER G, WEINZIERL M, NOLL C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers[J]. Clin Cancer Res, 2012, 18(6): 1619-1629.
|
| 24 |
ZHAN L Y, LEI S Q, LI W L, et al. Suppression of microRNA-142-5p attenuates hypoxia-induced apoptosis through targeting SIRT7[J]. Biomed Pharmacother, 2017, 94: 394-401.
|
| 25 |
WANG A, KWEE L C, GRASS E, et al. Whole blood sequencing reveals circulating microRNA associations with high-risk traits in non-ST-segment elevation acute coronary syndrome[J]. Atherosclerosis, 2017, 261: 19-25.
|
| 26 |
ZARJOU A, YANG S Z, ABRAHAM E, et al. Identification of a microRNA signature in renal fibrosis: role of miR-21[J]. Am J Physiol Renal Physiol, 2011, 301(4): F793-F801.
|
| 27 |
WANG H, WANG Z X, TANG Q B. Reduced expression of microRNA-199a-3p is associated with vascular endothelial cell injury induced by type 2 diabetes mellitus[J]. Exp Ther Med, 2018, 16(4): 3639-3645.
|
| 28 |
DENHAM N C, PEARMAN C M, CALDWELL J L, et al. Calcium in the pathophysiology of atrial fibrillation and heart failure[J]. Front Physiol, 2018, 9: 1380.
|
| 29 |
URBANTAT R M, VAJKOCZY P, BRANDENBURG S. Advances in chemokine signaling pathways as therapeutic targets in glioblastoma[J]. Cancers, 2021, 13(12): 2983.
|
| 30 |
LIU P, SUN H K, ZHOU X, et al. CXCL12/CXCR4 axis as a key mediator in atrial fibrillation via bioinformatics analysis and functional identification[J]. Cell Death Dis, 2021, 12(9): 813.
|
| 31 |
KAWASAKI M, MEULENDIJKS E R, VAN DEN BERG N W E, et al. Neutrophil degranulation interconnects over-represented biological processes in atrial fibrillation[J]. Sci Rep, 2021, 11(1): 2972.
|
| 32 |
ERTAS G, SÖNMEZ O, TURFAN M,et al. Neutrophil/lymphocyte ratio is associated with thromboembolic stroke in patients with non-valvular atrial fibrillation[J]. J Neurol Sci, 2013, 324(1/2): 49-52.
|
| 33 |
LI S B, YANG F, JING L, et al. Myeloperoxidase and risk of recurrence of atrial fibrillation after catheter ablation[J]. J Investig Med, 2013, 61(4): 722-727.
|
 )
)
 )
)