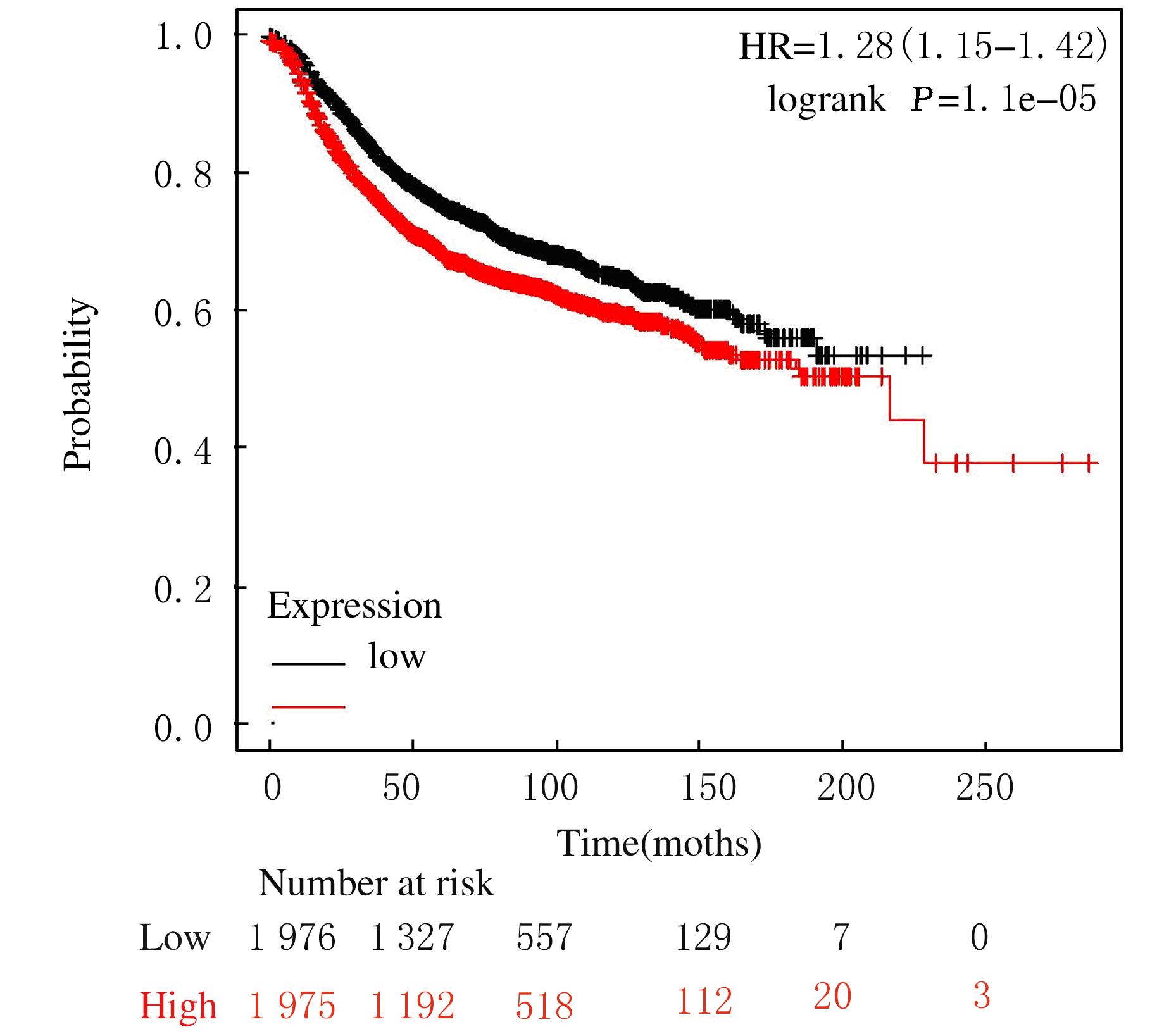
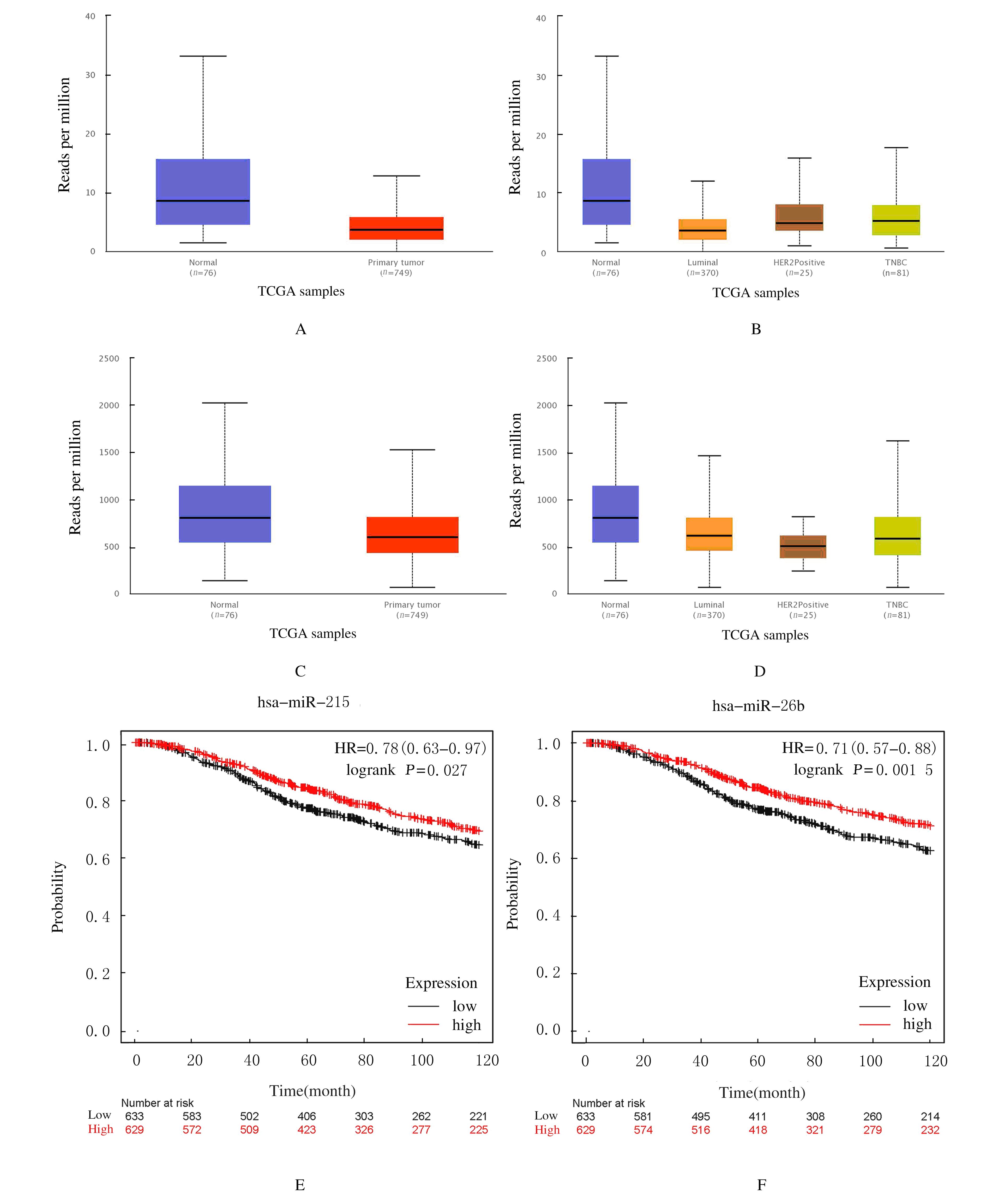
Journal of Jilin University(Medicine Edition) ›› 2021, Vol. 47 ›› Issue (5): 1229-1236.doi: 10.13481/j.1671-587X.20210521
• Research in clinical medicine • Previous Articles Next Articles
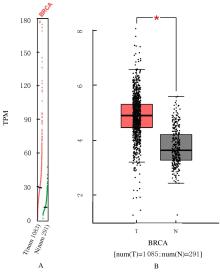
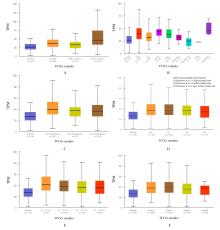
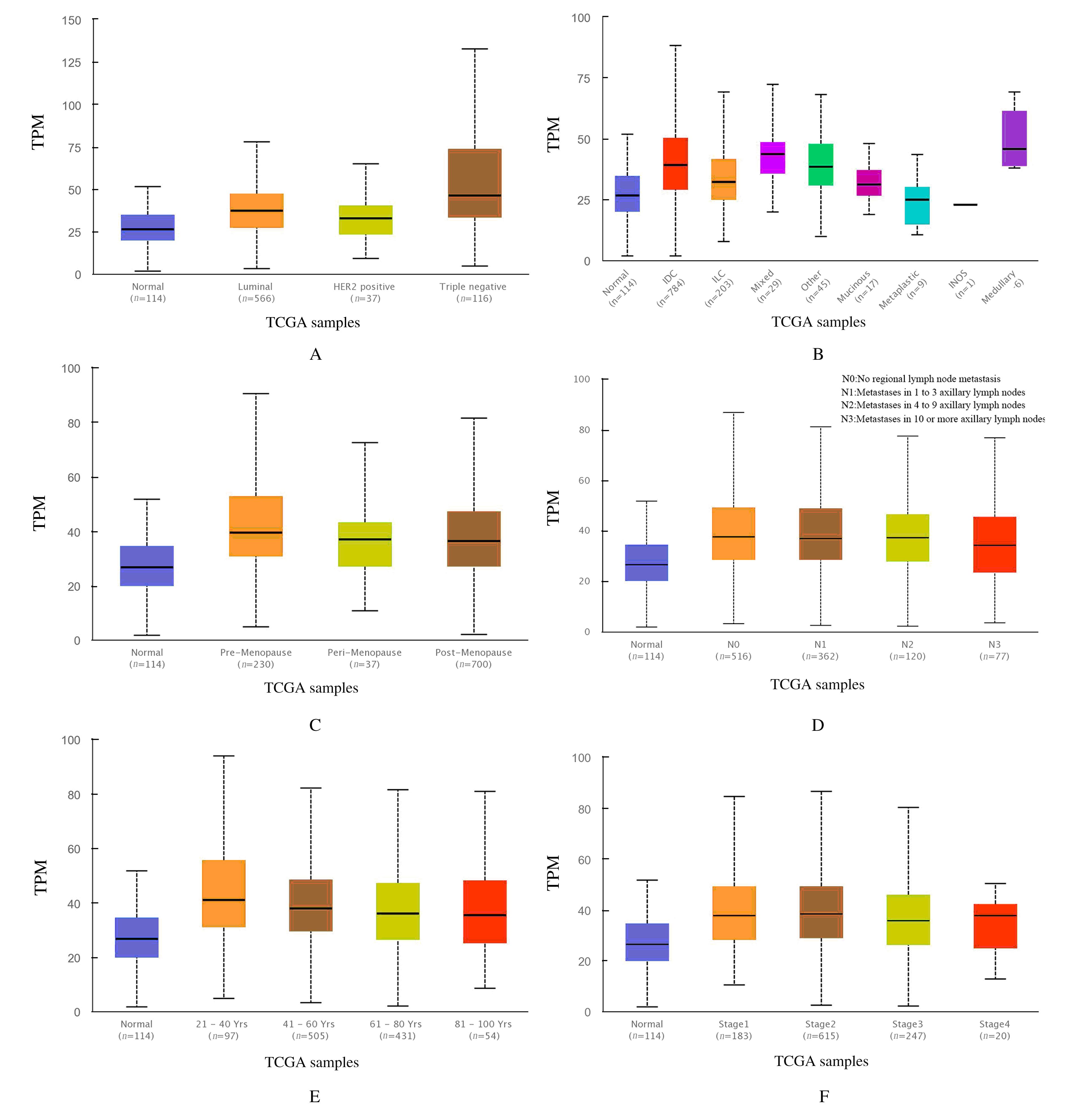
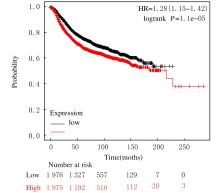
Bioinformatic analysis on expression characteristics of PRPS2 and its relationship with prognosis of breast cancer
Yang YU1,Sainan LIU2,Yunkai LIU1,Yong LI2,Yichun QIAO2,Yi CHENG1( )
)
- 1.Institute of Translational Medicine,First Hospital,Jilin University,Changchun 130021,China
2.Department of Epidemiology and Health Statistics,School of Public Health,Jilin University,Changchun 130021,China
-
Received:2021-02-12Online:2021-09-28Published:2021-10-26 -
Contact:Yi CHENG E-mail:chengyi@jlu.edu.cn
CLC Number:
- Q71
Cite this article
Yang YU,Sainan LIU,Yunkai LIU,Yong LI,Yichun QIAO,Yi CHENG. Bioinformatic analysis on expression characteristics of PRPS2 and its relationship with prognosis of breast cancer[J].Journal of Jilin University(Medicine Edition), 2021, 47(5): 1229-1236.
share this article
Tab.1
Top 25 co-expression genes of PRPS2"
| Correlated gene | Cytoband | Spearman’s correlation | P value | q value |
|---|---|---|---|---|
| CCSAP | 1q42.13 | 0.402 | 1.46E-38 | 2.95E-34 |
| POLA1 | Xp22.11-p21.3 | 0.396 | 1.87E-37 | 1.88E-33 |
| NKRF | Xq24 | 0.389 | 4.04E-36 | 2.47E-32 |
| TIPRL | 1q24.2 | 0.389 | 4.90E-36 | 2.47E-32 |
| FANCB | Xp22.2 | 0.381 | 1.54E-34 | 6.23E-31 |
| UBA2 | 19q13.11 | 0.374 | 3.17E-33 | 1.07E-29 |
| UCHL5 | 1q31.2 | 0.368 | 3.49E-32 | 1.01E-28 |
| MMS22L | 6q16.1 | 0.357 | 2.92E-30 | 6.54E-27 |
| GMPS | 3q25.31 | 0.356 | 4.22E-30 | 8.35E-27 |
| LIN9 | 1q42.12 | 0.356 | 4.93E-30 | 8.35E-27 |
| PIGA | Xp22.2 | 0.356 | 4.97E-30 | 8.35E-27 |
| HPRT1 | Xq26.2-q26.3 | 0.352 | 2.44E-29 | 3.68E-26 |
| C4ORF46 | 4q32.1 | 0.352 | 2.55E-29 | 3.68E-26 |
| SPIN4 | Xq11.1 | 0.351 | 3.27E-29 | 4.40E-26 |
| FAM72A | 1q32.1 | 0.350 | 4.72E-29 | 5.94E-26 |
| KIF14 | 1q32.1 | 0.349 | 6.16E-29 | 6.90E-26 |
| AHCTF1 | 1q44 | 0.348 | 8.64E-29 | 9.18E-26 |
| PPP1CB | 2p23.2 | 0.347 | 1.30E-28 | 1.32E-25 |
| CACYBP | 1q25.1 | 0.347 | 1.66E-28 | 1.60E-25 |
| HCCS | Xp22.2 | 0.346 | 2.37E-28 | 2.18E-25 |
| DEPDC1 | 1p31.3 | 0.344 | 3.94E-28 | 3.31E-25 |
| ZNF124 | 1q44 | 0.344 | 4.96E-28 | 4.01E-25 |
| ACTL6A | 3q26.33 | 0.344 | 5.49E-28 | 4.26E-25 |
| TTK | 6q14.1 | 0.343 | 6.57E-28 | 4.90E-25 |
| DKC1 | Xq28 | 0.342 | 8.88E-28 | 6.39E-25 |
| 1 | LI H, YAN M, WU X, et al. Expression and clinical significance of pyruvate kinase M2 in breast cancer: A protocol for meta-analysis and bioinformatics validation analysis [J]. Medicine, 2021, 100(18): e25545. |
| 2 | TANG Y, WANG Y, KIANI M F, et al. Classification, treatment strategy, and associated drug resistance in breast cancer [J]. Clin Breast Cancer, 2016, 16(5): 335-43. |
| 3 | PEDLEY A M, BENKOVIC S J. A new view into the regulation of purine metabolism: The purinosome [J]. Trends in biochemical sciences, 2017, 42(2): 141-154. |
| 4 | BECKER M A, AHMED M. Cell type-specific differential expression of human PRPP synthetase (PRPS) genes [J]. Adv Exp Med Biol, 2000, 486: 5-10. |
| 5 | CUNNINGHAM J T, MORENO M V, LODI A, et al. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer [J]. Cell, 2014, 157(5): 1088-103. |
| 6 | LV Y, WANG X, LI X, et al. Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway [J]. PLoS Biol, 2020, 18(11): e3000872. |
| 7 | TANG Z, LI C, KANG B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses [J].Nucleic Acids Res,2017,45(W1): W98-W102. |
| 8 | GYORFFY B, LANCZKY A, EKLUND A C, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients [J]. Breast Cancer Res Treat, 2010, 123(3): 725-731. |
| 9 | CERAMI E, GAO J, DOGRUSOZ U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data [J]. Cancer Discov, 2012, 2(5): 401-404. |
| 10 | HSU S D, LIN F M, WU W Y, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions [J].Nucleic Acids Res,2011,39(Database issue): D163-D169. |
| 11 | HUANG H Y, LIN Y C, LI J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database [J]. Nucleic Acids Res, 2020, 48(D1): D148-D154. |
| 12 | TONG X, ZHAO F, THOMPSON C B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells [J]. Curr Opin Genet Dev, 2009, 19(1): 32-37. |
| 13 | MANNAVA S, GRACHTCHOUK V, WHEELER L J,et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells [J]. Cell Cycle, 2008, 7(15): 2392-2400. |
| 14 | BURKHART R A, PINEDA D M, CHAND S N,et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer [J]. RNA Biol, 2013, 10(8): 1312-1323. |
| 15 | MIAO W, WANG Y. Targeted quantitative kinome analysis identifies PRPS2 as a promoter for colorectal cancer metastasis [J]. J Proteome Res, 2019, 18(5): 2279-2286. |
| 16 | LI T, SONG L, ZHANG Y, et al. Molecular mechanism of c-Myc and PRPS1/2 against thiopurine resistance in Burkitt’s lymphoma [J]. J Cell Mol Med, 2020, 24(12): 6704-6715. |
| 17 | VISHNOI A, RANI S. MiRNA Biogenesis and Regulation of Diseases: An Overview [J]. Methods Mol Biol, 2017, 1509: 1-10. |
| 18 | WILKE C M, HESS J, KLYMENKO S V, et al. Expression of miRNA-26b-5p and its target TRPS1 is associated with radiation exposure in post-Chernobyl breast cancer [J]. Int J Cancer, 2018, 142(3): 573-583. |
| 19 | JING S, TIAN J, ZHANG Y, et al. Identification of a new pseudogenes/lncRNAs-hsa-miR-26b-5p-COL12A1 competing endogenous RNA network associated with prognosis of pancreatic cancer using bioinformatics analysis [J].Aging(Albany NY),2020,12(19):19107-19128. |
| 20 | CUI X W, QIAN Z L, LI C, et al. Identification of miRNA and mRNA expression profiles by PCR microarray in hepatitis B virusassociated hepatocellular carcinoma [J]. Mol Med Rep, 2018, 18(6): 5123-5132. |
| 21 | CHENG C C, YANG B L, CHEN W C, et al. STAT3 mediated miR-30a-5p inhibition enhances proliferation and inhibits apoptosis in colorectal cancer cells [J]. Int J Mol Sci, 2020, 21(19):7315. |
| 22 | MOKHLESI A, TALKHABI M. Comprehensive transcriptomic analysis identifies novel regulators of lung adenocarcinoma [J].J Cell Commun Signal,2020,14(4): 453-465. |
| 23 | GADEWAL N, KUMAR R, AHER S, et al. miRNA-mRNA profiling reveals prognostic impact of SMC1A expression in acute myeloid leukemia [J]. Oncol Res, 2020, 28(3): 321-330. |
| [1] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
| [2] | Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765. |
| [3] | Shan GAO,Yutong WANG,Minqiu LU,Lei SHI,Bin CHU,Yuehua DING,Mengzhen WANG,Li BAO. Analysis on causes for early death and its risk factors of patients with multiple myeloma in era of novel drugs [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 783-789. |
| [4] | Chengsheng LI,Qihan BAO,Xiaoyan HAO,Qingzhong PAN,Suzhen WANG,Fuyan SHI. Establishment of prediction model for postoperative pancreatic cancer based on random forest algorithm [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 426-435. |
| [5] | Xiaoyan LI,Wei ZHANG,Jie HE. Promotion effect of REG1A on proliferation and migration of lung adenocarcinoma cells by regulating Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 444-453. |
| [6] | Ying ZHAO,Danyu ZHAO,Chao LIU. Bioinformatics analysis on prognostic evaluation value of TXNDC11 gene in pan-cancer and its immunity regulation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 142-153. |
| [7] | Zhijuan ZHAO,Lian MENG,Chunxia LIU. Bioinformatics analysis on miRNA-mRNA regulatory networks based on fusion genes acting in rhabdomyosarcoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 154-162. |
| [8] | Lili QIN,Xiaobo MA,Tianye ZHAO,Xuerong TAO,Min ZHENG,Xueying WANG,Jiaxin YI,Yanhua WU,Jing JIANG. Effects of MMP-9 and TIMP-1 expressions on prognostic evaluation of gastric cancer patients after radical gastrectomy [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 163-171. |
| [9] | Nan LI,Lei CHEN,Tianmin XU,Kun ZHANG. Bioinformatics analysis on screening of extracellular matrix related genes in patients with endometriosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 188-194. |
| [10] | Xiuyan YU,Ting LI,Zhanjie CONG,Wenlong WANG,Xiaowei ZHANG,Xuefeng WU. Combined detection of hMAM, SBEM and CEACAM19 mRNA in peripheral blood of breast cancer patients and its clinical significance [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 195-202. |
| [11] | Hao DONG,Haizhen JIANG,Chao LI,Dengke GAO,Yaping JIN,Huatao CHEN. Construction of mouse NR1D1 gene overexpression vector and its bioinformatics analysis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 94-103. |
| [12] | Shuzhen LI,Yajie CAO,Haiying GENG,Zengxiaorui CAI,Chunmei DAI,Youfeng WEN,Ning LI. Bioinformatics analysis based on expression level of HMMR in lung adenocarcinoma tissue and its impact on prognosis of LUAD patients [J]. Journal of Jilin University(Medicine Edition), 2021, 47(6): 1502-1509. |
| [13] | Xiuyan YU,Chunying TIAN,Xiaowei ZHANG,Xuefeng WU. Clinical significances of serum levels of SBEM, hMAM and CEACAM19 in early diagnosis of breast cancer [J]. Journal of Jilin University(Medicine Edition), 2021, 47(6): 1518-1525. |
| [14] | Aiping WEN,Le LUO,Zhiwei MENG,Jingji JIN,Honggui ZHOU,Jihong ZHU. Expression of Tip60 protein in endometrial adenocarcinoma tissue and its clinical significance [J]. Journal of Jilin University(Medicine Edition), 2021, 47(5): 1244-1249. |
| [15] | Yuqing PAN, Yunyan SUN, Yixun LI, Liying WANG, Zhuoma SINAN, Rui CHEN, Xi ZHANG, Yan DU. Construction and analysis of competitive endogenous RNA networks in acute myeloid leukemia based on high-throughput microarray [J]. Journal of Jilin University(Medicine Edition), 2021, 47(3): 669-676. |