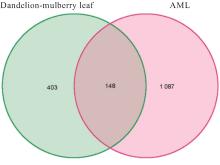
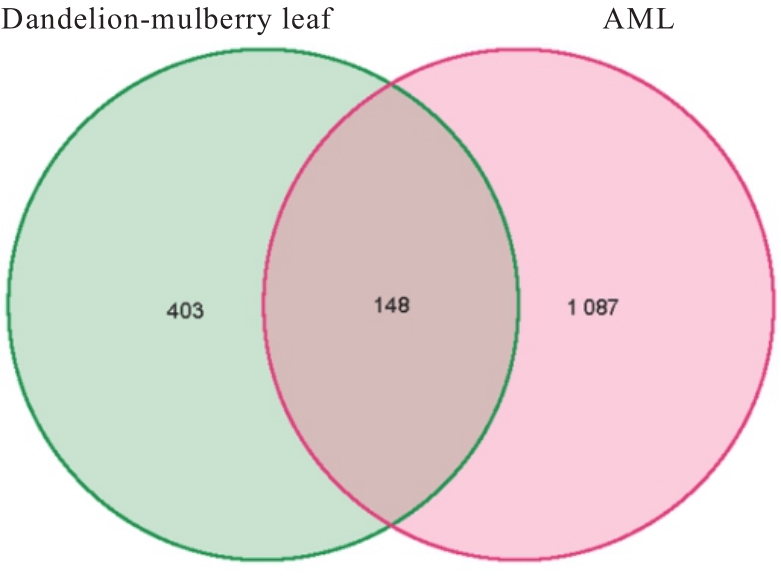
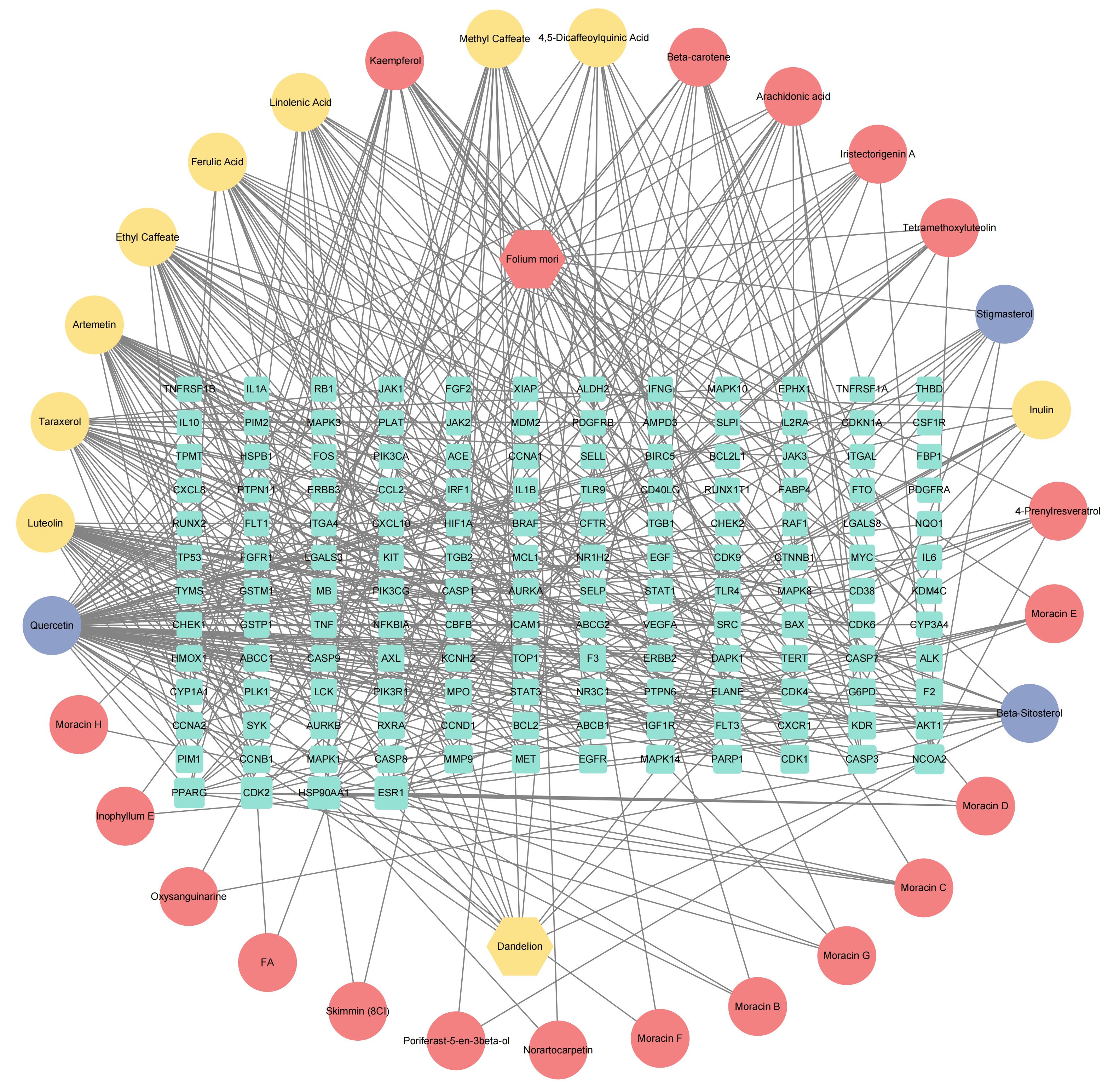
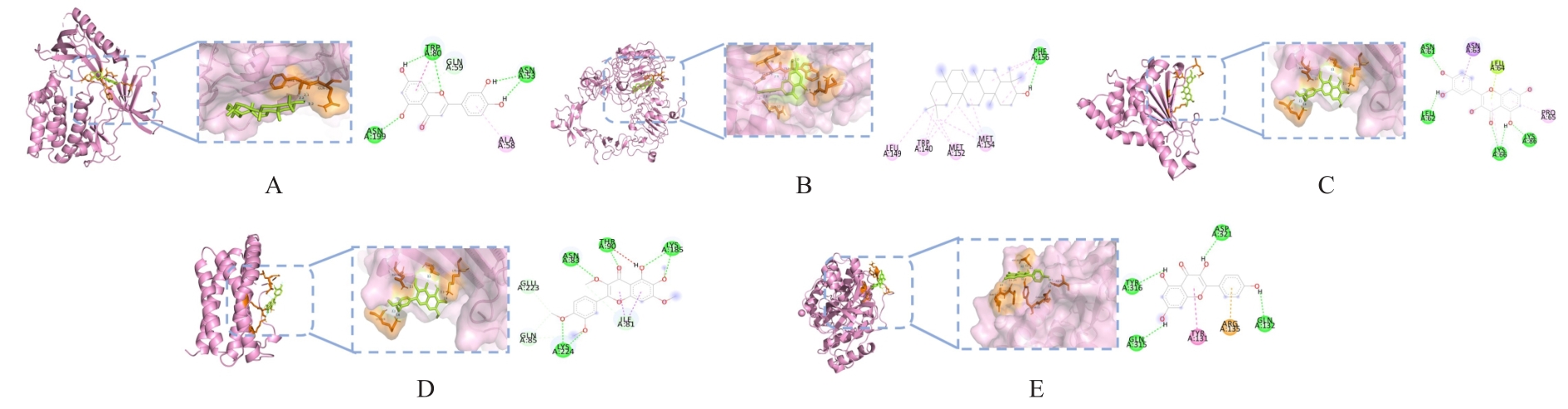
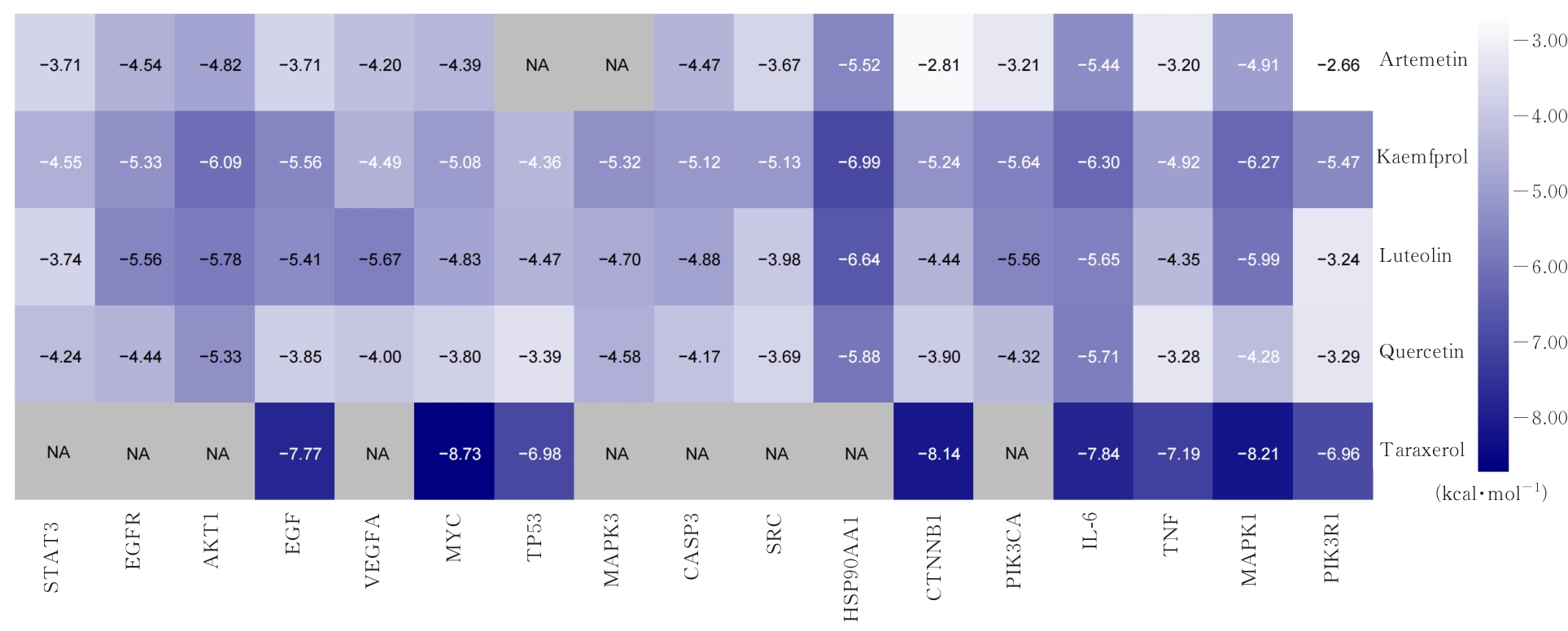
Journal of Jilin University(Medicine Edition) ›› 2024, Vol. 50 ›› Issue (4): 1087-1097.doi: 10.13481/j.1671-587X.20240423
• Research in clinical medicine • Previous Articles Next Articles
Network pharmacology analysis based on potential mechanism of dandelion-mulberry leaf in treatment of acute myeloid leukemia
Xinchen ZHOU,Shuhan DONG,Zhuo ZHANG,Mingmei SHEN,Xiangjun WANG,Ying LI,Limei LIU( )
)
- Department of Microbiology Laboratory,School of Medical Technology,Beihua University,Jilin 132000,China
-
Received:2023-12-16Online:2024-07-28Published:2024-08-01 -
Contact:Limei LIU E-mail:liulm74@163.com
CLC Number:
- R733.7
Cite this article
Xinchen ZHOU,Shuhan DONG,Zhuo ZHANG,Mingmei SHEN,Xiangjun WANG,Ying LI,Limei LIU. Network pharmacology analysis based on potential mechanism of dandelion-mulberry leaf in treatment of acute myeloid leukemia[J].Journal of Jilin University(Medicine Edition), 2024, 50(4): 1087-1097.
share this article
| 1 | SIEGEL R L, MILLER K D, FUCHS H E,et al.Cancer statistics, 2022[J]. CA: Cancer J Clin,2022, 72 (1): 7-33. |
| 2 | 梁欣荃, 唐亦舒, 朱 平, 等. 不同类型急性白血病患者血流感染流行病学及预后分析: 一项长达九年多中心947例患者回顾性研究[J]. 临床血液学杂志, 2023, 36(1): 27-32. |
| 3 | NAGEL G, WEBER D, FROMM E, et al. Epidemiological, genetic, and clinical characterization by age of newly diagnosed acute myeloid leukemia based on an academic population-based registry study (AMLSG BiO)[J]. Ann Hematol, 2017, 96(12): 1993-2003. |
| 4 | ZENG Y J, WU M, ZHANG H, et al. Effects of qinghuang powder on acute myeloid leukemia based on network pharmacology, molecular docking, and in vitro experiments[J]. Evid Based Complement Alternat Med, 2021, 2021: 6195174. |
| 5 | FANG T T, LIU L Q, LIU W J. Network pharmacology-based strategy for predicting therapy targets of Tripterygium wilfordii on acute myeloid leukemia[J]. Medicine (Baltimore), 2020, 99(50): e23546. |
| 6 | 霍俊明, 于维贤, 李维奇, 等. 急性白血病临床治疗体会与分析[J]. 中医杂志, 1987, 28(8): 31-33. |
| 7 | DEEPA M, SURESHKUMAR T, SATHEESHKUMAR P K, et al. Purified mulberry leaf lectin (MLL) induces apoptosis and cell cycle arrest in human breast cancer and colon cancer cells[J]. Chem Biol Interact, 2012, 200(1): 38-44. |
| 8 | 杨子辉, 董 朕, 伍蕙岚, 等. 基于网络药理学分析蒲公英抗氧化功能的物质基础与作用机制[J]. 畜牧兽医学报, 2023, 54(5): 2170-2185. |
| 9 | WU Z Y, JI X W, SHAN C, et al. Exploring the pharmacological components and effective mechanism of Mori Folium against periodontitis using network pharmacology and molecular docking[J]. Arch Oral Biol, 2022, 139: 105391. |
| 10 | RU J L, LI P, WANG J N, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines[J]. J Cheminform, 2014, 6: 13. |
| 11 | WU Y, ZHANG F L, YANG K, et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping[J]. Nucleic Acids Res, 2019, 47(D1): D1110-D1117. |
| 12 | LIU Z Y, GUO F F, WANG Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine[J]. Sci Rep, 2016, 6: 21146. |
| 13 | UNIPROT CONSORTIUM. UniProt: a worldwide hub of protein knowledge[J]. Nucleic Acids Res, 2019, 47(D1): D506-D515. |
| 14 | PIÑERO J, RAMÍREZ-ANGUITA J M, SAÜCH-PITARCH J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update[J]. Nucleic Acids Res, 2020, 48(D1): D845-D855. |
| 15 |
STELZER G, ROSEN N, PLASCHKES I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses[J]. Curr Protoc Bioinformatics, 2016, 54. DOI: 10.1002/cpbi.5 .
doi: 10.1002/cpbi.5 |
| 16 | AMBERGER J S, HAMOSH A. Searching online Mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes[J]. Curr Protoc Bioinformatics, 2017, 58: 1.2.1-1.2.1.2.12. |
| 17 | SZKLARCZYK D, GABLE A L, NASTOU K C, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets[J]. Nucleic Acids Res, 2021, 49(D1): D605-D612. |
| 18 | GONZÁLEZ-CASTEJÓN M, VISIOLI F, RODRIGUEZ-CASADO A. Diverse biological activities of dandelion[J]. Nutr Rev, 2012, 70(9): 534-547. |
| 19 | ZHANG Y L, LIANG Y E, HE C W. Anticancer activities and mechanisms of heat-clearing and detoxicating traditional Chinese herbal medicine[J]. Chin Med, 2017, 12: 20. |
| 20 | MA G Q, CHAI X Y, HOU G G, et al. Phytochemistry, bioactivities and future prospects of mulberry leaves: a review[J]. Food Chem, 2022, 372: 131335. |
| 21 | 刘晓燕, 龙 凤, 赵 玉, 等. 蒲公英中有效成分抗肿瘤作用机制的研究进展[J]. 中草药, 2023, 54(10): 3391-3400. |
| 22 | 袁鑫怡, 曾淑欣, 杨 润, 等. 基于网络药理学与分子对接的蒲公英抗乳腺癌的机制研究[J]. 天津中医药, 2023, 40(1): 110-116. |
| 23 | OVADJE P, CHATTERJEE S, GRIFFIN C, et al. Selective induction of apoptosis through activation of caspase-8 in human leukemia cells (Jurkat) by dandelion root extract[J]. J Ethnopharmacol, 2011, 133(1): 86-91. |
| 24 | YU H, PARDOLL D, JOVE R. STATs in cancer inflammation and immunity: a leading role for STAT3[J]. Nat Rev Cancer, 2009, 9(11): 798-809. |
| 25 | LARRUE C, HEYDT Q, SALAND E, et al. Oncogenic KIT mutations induce STAT3-dependent autophagy to support cell proliferation in acute myeloid leukemia[J]. Oncogenesis, 2019, 8: 39. |
| 26 | VENUGOPAL S, BAR-NATAN M, MASCARENHAS J O. JAKs to STATs: a tantalizing therapeutic target in acute myeloid leukemia[J]. Blood Rev, 2020, 40: 100634. |
| 27 | ZENG Y J, LIU F, WU M, et al. Curcumin combined with arsenic trioxide in the treatment of acute myeloid leukemia: network pharmacology analysis and experimental validation[J]. J Cancer Res Clin Oncol, 2023, 149(1): 219-230. |
| 28 | AMJAD E, SOKOUTI B, ASNAASHARI S. A systematic review of anti-cancer roles and mechanisms of kaempferol as a natural compound[J]. Cancer Cell Int, 2022, 22(1): 260. |
| 29 | XIAO J, ZHANG B, YIN S M, et al. Quercetin induces autophagy-associated death in HL-60 cells through CaMKKβ/AMPK/mTOR signal pathway[J]. Acta Biochim Biophys Sin, 2022, 54(9): 1244-1256. |
| 30 | SHAHBAZ M, IMRAN M, ALSAGABY S A, et al. Anticancer, antioxidant, ameliorative and therapeutic properties of kaempferol[J]. Int J Food Prop, 2023, 26(1): 1140-1166. |
| 31 | Al-Mosawe E H.The effect of luteolin on lymphocyte cells in leukemia patient[J]. Al-Mustansiriyah J Sci, 2013, 24 (1): 1-8. |
| 32 | ZHENG D D, ZHOU Y M, LIU Y, et al. Molecular mechanism investigation on monomer kaempferol of the traditional medicine Dingqing Tablet in promoting apoptosis of acute myeloid leukemia HL-60 cells[J]. Evid Based Complement Alternat Med, 2022, 2022: 8383315. |
| 33 | MUS A A, GOH L P W, MARBAWI H, et al. The biosynthesis and medicinal properties of taraxerol[J]. Biomedicines, 2022, 10(4): 807. |
| 34 | KHANRA R, DEWANJEE S, DUA T K, et al. Taraxerol, a pentacyclic triterpene from Abroma Augusta leaf, attenuates acute inflammation via inhibition of NF-κB signaling[J]. Biomedecine Pharmacother, 2017, 88: 918-923. |
| 35 | KHANRA R, BHATTACHARJEE N, DUA T K, et al. Taraxerol, a pentacyclic triterpenoid, from Abroma Augusta leaf attenuates diabetic nephropathy in type 2 diabetic rats[J]. Biomedecine Pharmacother, 2017, 94: 726-741. |
| 36 | YAO X Y, LI G L, BAI Q, et al. Taraxerol inhibits LPS-induced inflammatory responses through suppression of TAK1 and Akt activation[J]. Int Immunopharmacol, 2013, 15(2): 316-324. |
| 37 | YAOI X, LU B Y, LÜ C T, et al. Taraxerol induces cell apoptosis through A mitochondria-mediated pathway in HeLa cells[J]. Cell J, 2017, 19(3): 512-519. |
| 38 | KO W G, KANG T H, LEE S J, et al. Polymethoxyflavonoids from Vitex rotundifolia inhibit proliferation by inducing apoptosis in human myeloid leukemia cells[J]. Food Chem Toxicol, 2000, 38(10): 861-865. |
| 39 | MARTELLI A M, NYÅKERN M, TABELLINI G, et al. Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia[J]. Leukemia, 2006, 20(6): 911-928. |
| [1] | Shan CAO,Yijia ZHANG,Yang BAI,Fang CHEN,Sha XIE,Qianqian HAN. Network pharmacological analysis and in vitro experimental verification based on anti-atherosclerosis mechanism of Xiaoban Tongmai Formula [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 925-938. |
| [2] | Jinli TAN,Dan HUANG,Jingyang LIAO,Liuchong ZHU,Wenbin LIU. Molecular docking analysis on screening of novel antibacterial targets and their drugs of Staphylococcus aureus based on pan-genomics and subtractive proteomics techniques [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 970-977. |
| [3] | Zijia ZHU,Xia CHEN,Man CUI,Jihong WEN,Ping WANG,Dong SONG. Bioinformatics and molecular docking technology analysis on mechanism of salidroside on key differential genes of triple negative breast cancer [J]. Journal of Jilin University(Medicine Edition), 2024, 50(3): 759-769. |
| [4] | Yang YU,Dan TIAN,Donghe NI,Duo ZHANG. Network pharmacologry and molecular docking analysis based on mechanism of monk fruit in treatment of diabetic nephropathy [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 161-167. |
| [5] | Aiying CHEN,Jinwen JIANG,Hui ZHANG. Network pharmacology and molecular docking analysis based on mechanism of Huangqin Tang in treatment of colorectal cancer [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 208-220. |
| [6] | Li LIU,Linsheng HUANG,Yongheng ZHAO,Wenjie CAO,Yongshuai QIAN,Huifan YU,Fei LI. Network pharmacological analysis on Balanophora involucrataHook.f. in treatment of hyperuricemia and its therapeutic effect on hyperuricemia cell model and hyperuricemia model mouse [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 58-70. |
| [7] | Zhongxin FENG,Mei LI. Effect of soluble CD40 ligand on biological behavior of THP-1 cells through long non-coding RNA linc00239 [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 88-96. |
| [8] | Chunling WANG,Sinuo WU,Xiaoyan YU,Weidong ZHANG. Analysis on network pharmacology and molecular docking technique based on effective chemical components and action targets of epimedium in treatment of hypothalamus-pituitary- adrenal gland/ gonad / thyroid gland axis function damage [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1296-1303. |
| [9] | Yumeng LIU,Song LENG, SARENGAOWA,Daijie LIN,Linsheng XIE,Mengrui LI,Xiao XU,Wannan LI. Network pharmacology analysis based on therapeutic effect of Sanghuang on pneumonia and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 923-930. |
| [10] | Sihan LAI,Juntong LIU,Luying TAN,Jinping LIU,Pingya LI. Network pharmacology and molecular docking analysis on anti-ischemic stroke mechanism of Panax quinquefolium triolsaponins [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 913-922. |
| [11] | Han WANG,Yali ZHANG,Haiwei QUAN,Runhong MU,Yutong ZOU,Wei XIA. Expression of miR-92a-3p in exosome in serum of patients with acute myeloid leukemia and its significance [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 467-472. |
| [12] | Haiwei QUAN,Yali ZHANG,Han WANG,Yijiu AI,Yutong ZOU,Wei XIA. Detection of expression level of miR-4286 in exosome in peripheral blood of patients with acute myeloid leukemia and its diagnostic value [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 460-466. |
| [13] | Zhouquan LI,Hui LI,Ying TANG,Lijun YANG,Yinghong JIANG,Lipin YIN. Network pharmacology and molecular docking analysis on mechanism of Tongluo Tangtai Power in treatment of diabetic peripheral neuropathy [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 341-350. |
| [14] | Yushuang GONG,Yefan FU,Huijing XU,Jian GUO,Lin XIANG,Rui HU,Rui FAN,Jie WANG,Miao LI,Meiyan SUN. Network pharmacology analysis on mechanism of Tonifying Slpeen and Kidney Empirical Prescription in treatment of neuromyelitis optica spectrum disorders [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 261-271. |
| [15] | Minghui WANG,Moyi LIU,Helin WANG,Ying LI,Xiangjun WANG,Hetong HUI,Xinyuan FAN,Tianqi WANG,Limei LIU. Bioinformatics analysis of network pharmacology and molecular docking technology based on mechanism of Physalis Calyx seu Fructus on leukemia [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 74-83. |
|