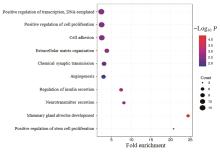
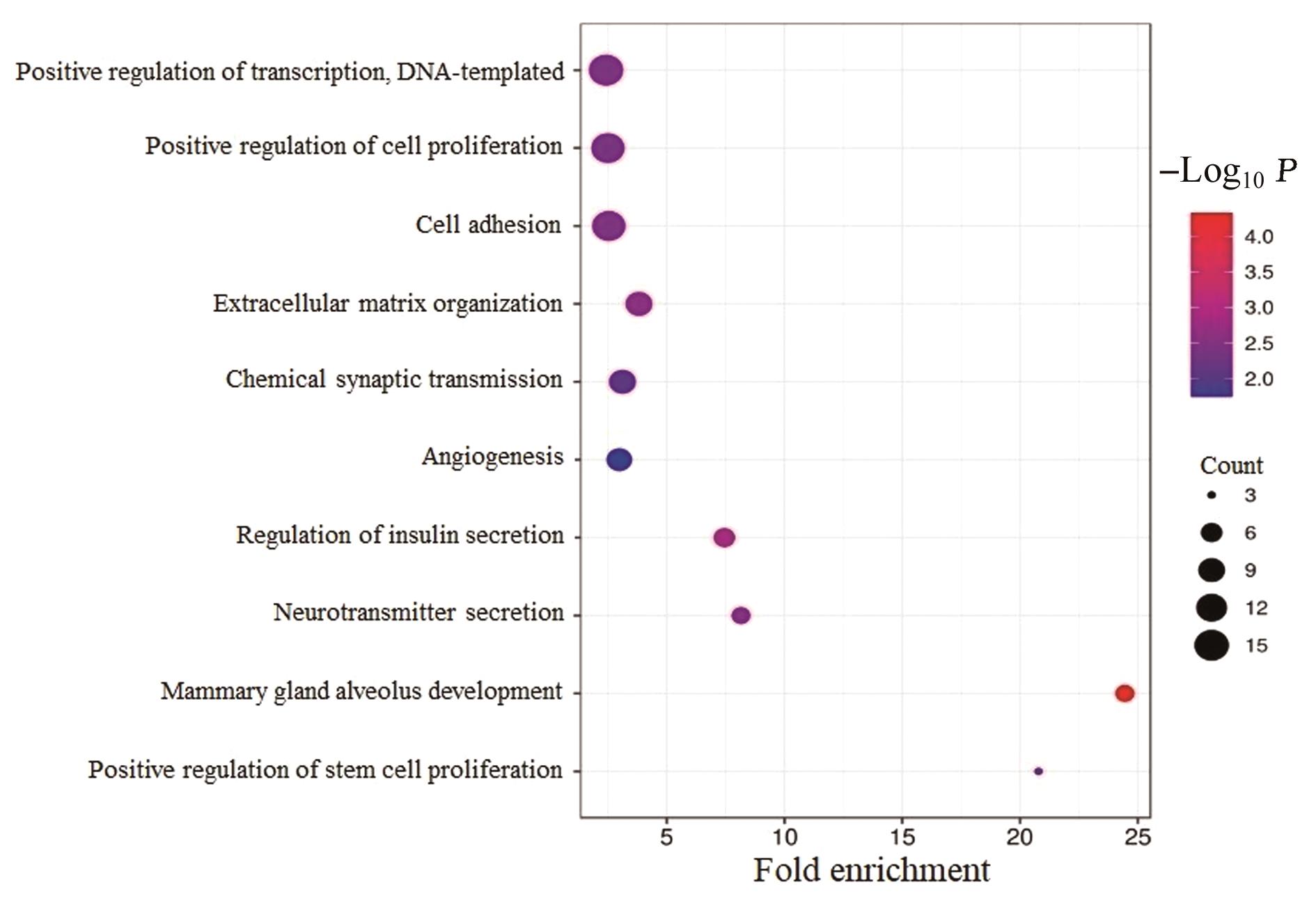
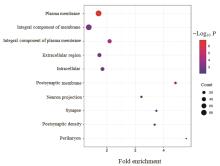
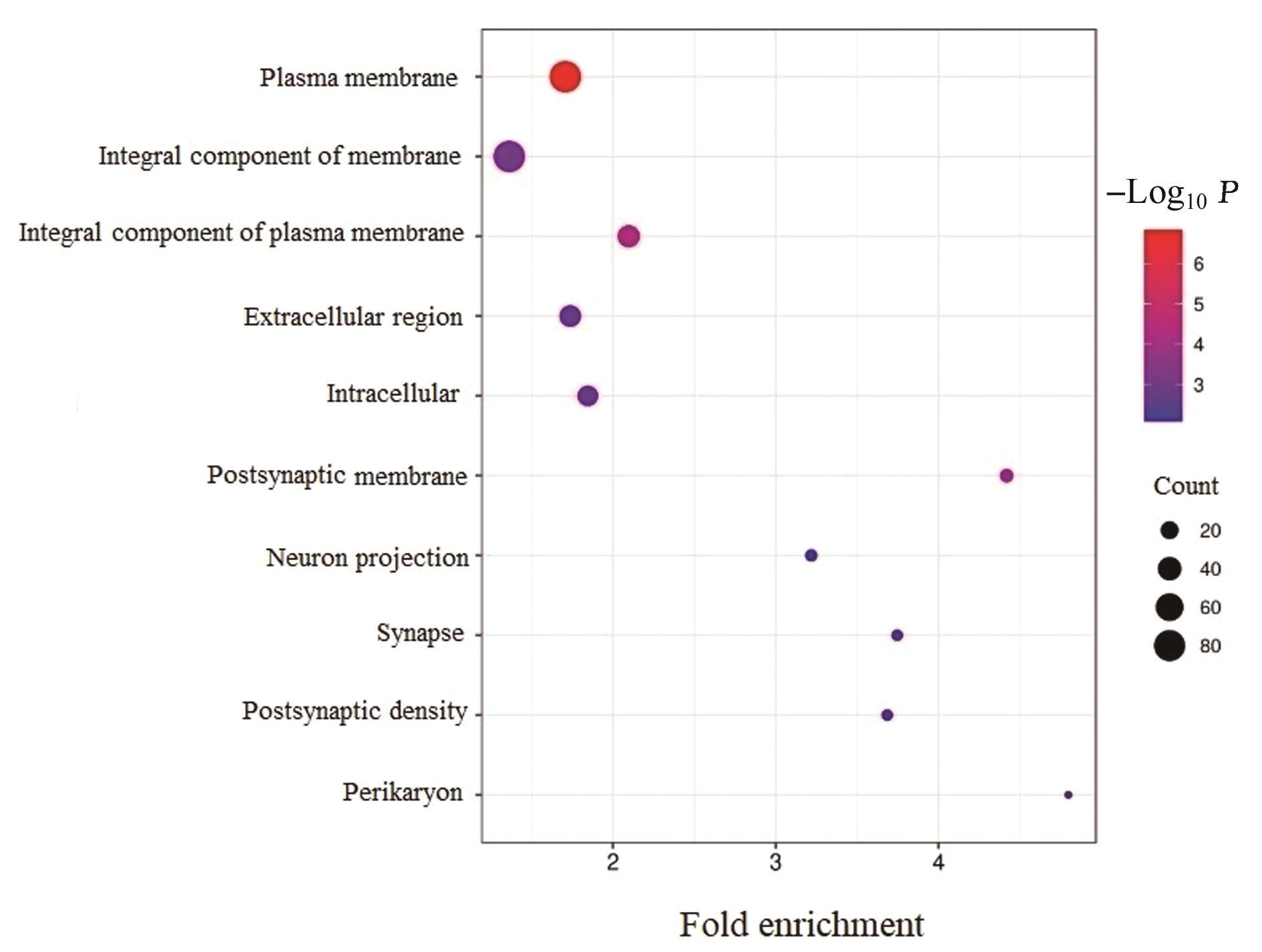
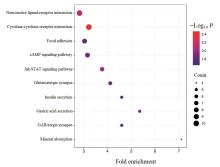
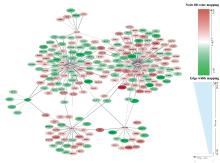
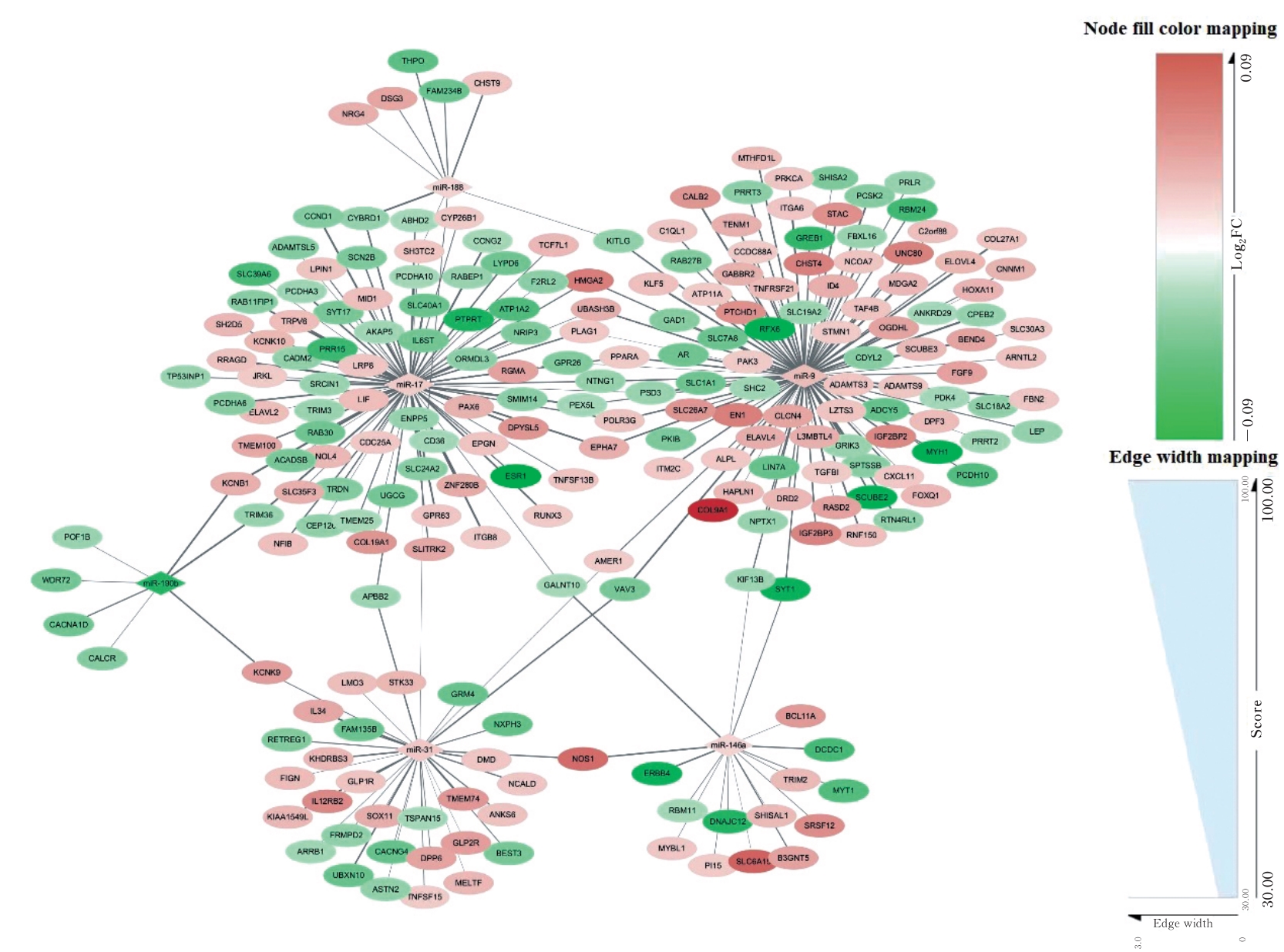
| 1 |
KWAN M L, BERNARD P S, KROENKE C H, et al. Breastfeeding, PAM50 tumor subtype, and breast cancer prognosis and survival[J].J Natl Cancer Inst, 2015, 107(7): djv087.
|
| 2 |
YUAN Z Y, WANG S S, GAO Y, et al. Clinical characteristics and prognosis of triple-negative breast cancer: a report of 305 cases[J].Ai Zheng,2008,27(6): 561-565.
|
| 3 |
EBERLEIN T J. Race, breast cancer subtypes, and survival in the Carolina breast cancer study[J]. Yearb Surg, 2007, 2007: 304-305.
|
| 4 |
NAKAJIMA H, FUJIWARA I, MIZUTA N, et al. Prognosis of Japanese breast cancer based on hormone receptor and HER2 expression determined by immunohistochemical staining[J]. World J Surg, 2008, 32(11): 2477-2482.
|
| 5 |
BABYSHKINA N, MALINOVSKAYA E, PATALYAK S, et al. Neoadjuvant chemotherapy for different molecular breast cancer subtypes: a retrospective study in Russian population[J]. Med Oncol, 2014, 31(9): 165.
|
| 6 |
GROUP I B C S, COLLEONI M, GELBER S, et al. Tamoxifen after adjuvant chemotherapy for premenopausal women with lymph node-positive breast cancer: international Breast Cancer Study Group Trial 13-93[J]. J Clin Oncol, 2006, 24(9): 1332-1341.
|
| 7 |
Straver M E, Rutgers E J T, Rodenhuis S,et al.The relevance of breast cancer subtypes in the outcome of neoadjuvant chemotherapy[J]. Annal surgical Oncol, 2010, 17(9):2411-2418.
|
| 8 |
RAKHAE A, EL-SAYEDM E, GREENA R, et al. Prognostic markers in triple-negative breast cancer[J]. Cancer, 2007, 109(1): 25-32.
|
| 9 |
XIONG B, LEI X F, ZHANG L, et al. miR-103 regulates triple negative breast cancer cells migration and invasion through targeting olfactomedin 4[J]. Biomed Pharmacother, 2017, 89: 1401-1408.
|
| 10 |
LIANG Z X, BIAN X H, SHIM H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer[J]. Biochem Biophys Res Commun, 2016, 477(3): 461-466.
|
| 11 |
LEE R C, FEINBAUM R L, AMBROS V. The C. elegans heterochronic gene Lin-4 encodes small RNAs with antisense complementarity to Lin-14[J]. Cell, 1993, 75(5): 843-854.
|
| 12 |
SANTOVITO D, MEZZETTI A, CIPOLLONE F. microRNAs and atherosclerosis: new actors for an old movie[J]. Nutr Metab Cardiovasc Dis, 2012, 22(11): 937-943.
|
| 13 |
RUPAIMOOLE R, CALIN G A, LOPEZ-BERESTEIN G, et al. miRNA deregulation in cancer cells and the tumor microenvironment[J]. Cancer Discov, 2016, 6(3): 235-246.
|
| 14 |
CHUA J H, ARMUGAM A, JEYASEELAN K. microRNAs: biogenesis, function and applications[J]. Curr Opin Mol Ther, 2009, 11(2): 189-199.
|
| 15 |
LING H, FABBRI M, CALIN G A. microRNAs and other non-coding RNAs as targets for anticancer drug development[J]. Nat Rev Drug Discov, 2013, 12(11): 847-865.
|
| 16 |
VOLINIA S, CALIN G A, LIU C G,et al.A microRNA expression signature of human solid tumors defines cancer gene targets[J]. Proc Natl Acad Sci U S A, 2006, 103(7): 2257-2261.
|
| 17 |
JANG M H, KIM H J, GWAK J M, et al. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer[J]. Hum Pathol, 2017, 68: 69-78.
|
| 18 |
FAN C N, LIU N, ZHENG D, et al. microRNA-206 inhibits metastasis of triple-negative breast cancer by targeting transmembrane 4 L6 family member 1[J]. Cancer Manag Res, 2019, 11: 6755-6764.
|
| 19 |
BARONI S, ROMERO-CORDOBA S, PLANTAMURA I, et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts[J]. Cell Death Dis, 2016, 7(7): e2312.
|
| 20 |
D'IPPOLITO E, PLANTAMURA I, BONGIOVANNI L,et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer[J]. Cancer Res, 2016,76(18): 5562-5572.
|
| 21 |
JANG M H, KIM H J, GWAK J M, et al. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer[J]. Hum Pathol, 2017, 68: 69-78.
|
| 22 |
D'IPPOLITO E, PLANTAMURA I, BONGIOVANNI L,et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer[J]. Cancer Res, 2016, 76(18): 5562-5572.
|
| 23 |
LI J, LAI Y H, MA J Y, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer[J]. BMC Cancer, 2017, 17(1): 745.
|
| 24 |
LUO L J, YANG F, DING J J, et al. miR-31 inhibits migration and invasion by targeting SATB2 in triple negative breast cancer[J]. Gene, 2016, 594(1): 47-58.
|
| 25 |
SI C S, YU Q, YAO Y F. Effect of miR-146a-5p on proliferation and metastasis of triple-negative breast cancer via regulation of SOX5[J]. Exp Ther Med, 2018, 15(5): 4515-4521.
|
| 26 |
ZAVALA V, PÉREZ-MORENO E, TAPIA T, et al. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival[J]. Cancer Biomark, 2016, 16(1): 99-107.
|
| 27 |
NAOREM L D, MUTHAIYAN M, VENKATESAN A. Identification of dysregulated miRNAs in triple negative breast cancer: a meta-analysis approach[J]. J Cell Physiol, 2019, 234(7): 11768-11779.
|
| 28 |
HAMILTON N, AUSTIN D, MÁRQUEZ-GARBÁN D, et al. Receptors for insulin-like growth factor-2 and androgens as therapeutic targets in triple-negative breast cancer[J]. Int J Mol Sci, 2017, 18(11): 2305.
|
| 29 |
HAMILTON N, AUSTIN D, MÁRQUEZ-GARBÁN D,et al. Receptors for insulin-like growth factor-2 and androgens as therapeutic targets in triple-negative breast cancer[J]. Int J Mol Sci, 2017, 18(11): 2305.
|
| 30 |
WANG Z S, LI Y F, XIAO Y J, et al. Integrin α9 depletion promotes β-catenin degradation to suppress triple-negative breast cancer tumor growth and metastasis[J]. Int J Cancer, 2019, 145(10): 2767-2780.
|
| 31 |
RIGIRACCIOLO D C, NOHATA N, LAPPANO R, et al. IGF-1/IGF-1R/FAK/YAP transduction signaling prompts growth effects in triple-negative breast cancer (TNBC) cells[J]. Cells, 2020, 9(4): 1010.
|
| 32 |
SHAHEEN S, FAWAZ F, SSHA H,et al. Differential expression and pathway analysis in drug-resistant triple-negative breast cancer cell lines using RNASeq analysis[J]. Int J Mol Sci,2018,19(6):1810.
|
| 33 |
WANG S, XIA D, WANG X,et al. C/EBPβ regulates the JAK/STAT signaling pathway in triple-negative breast cancer[J]. FEBS Open Bio, 2021, 11: 1250-1258.
|
 ),李卓林2(
),李卓林2( )
)
 ),Zhuolin LI2(
),Zhuolin LI2( )
)