吉林大学学报(医学版) ›› 2025, Vol. 51 ›› Issue (1): 115-123.doi: 10.13481/j.1671-587X.20250114
• 基础研究 • 上一篇
沉默DDX39A基因对食管癌TE-1细胞增殖、迁移和侵袭的作用及其机制
- 1.河北北方学院研究生学院,河北 张家口 075000
2.陆军第八十一集团军医院肿瘤内科,河北 张家口 075000
3.河北北方学院附属第一医院病理科,河北 张家口 075000
4.河北北方学院 基础医学院病理教研室,河北 张家口 075000
Effect of silencing DDX39A gene on proliferation, migration and invasion of esophageal cancer TE-1 cells and its mechanism
Pengli WU1,Fengyu LI2,Bo LIU3,Yang LYU4( )
)
- 1.Graduate School,Hebei North University,Zhangjiakou 075000,China
2.Department of Medical Oncology,Army 81st Group Army Hospital,Zhangjiakou 075000,China
3.Department of Pathology,First Affiliated Hospital,Hebei North University,Zhangjiakou 075000,China
4.Department of Pathology,Hebei North University,Zhangjiakou 075000,China
摘要:
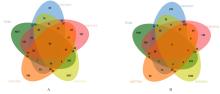
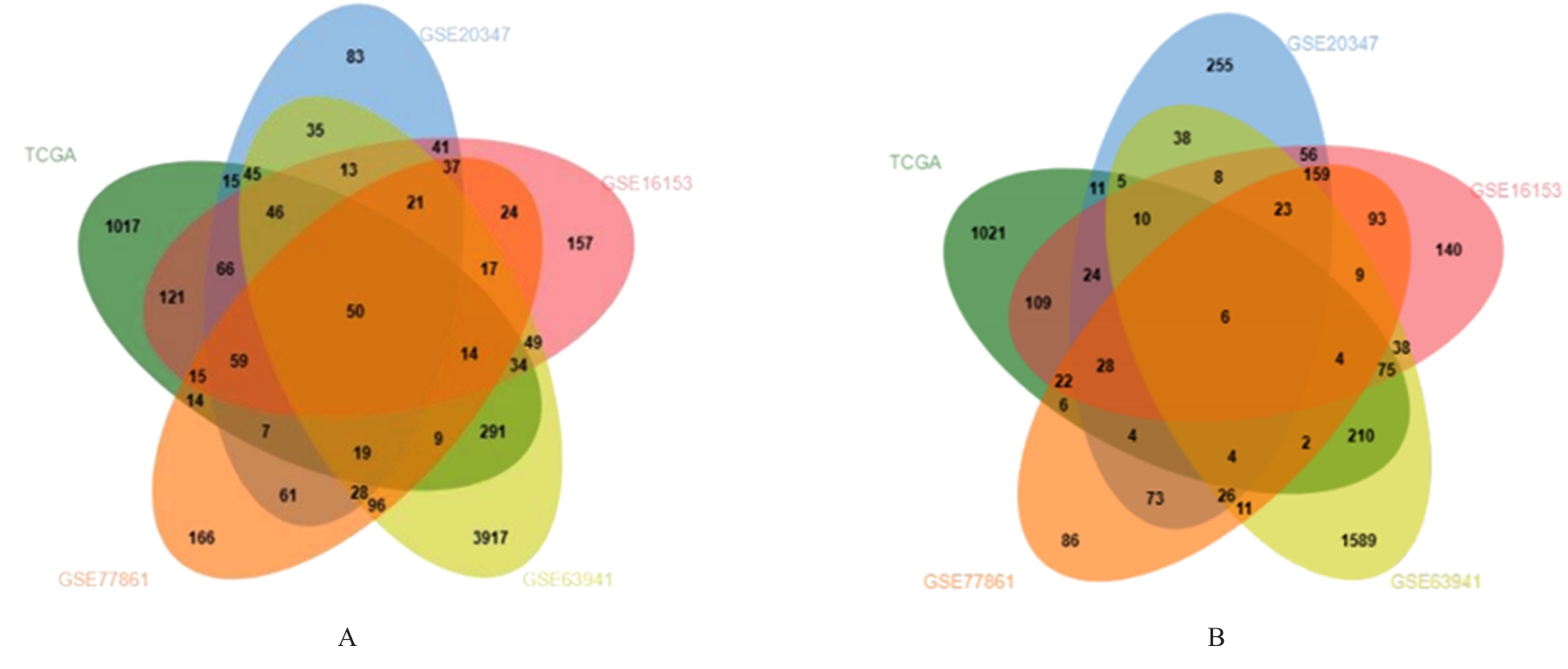
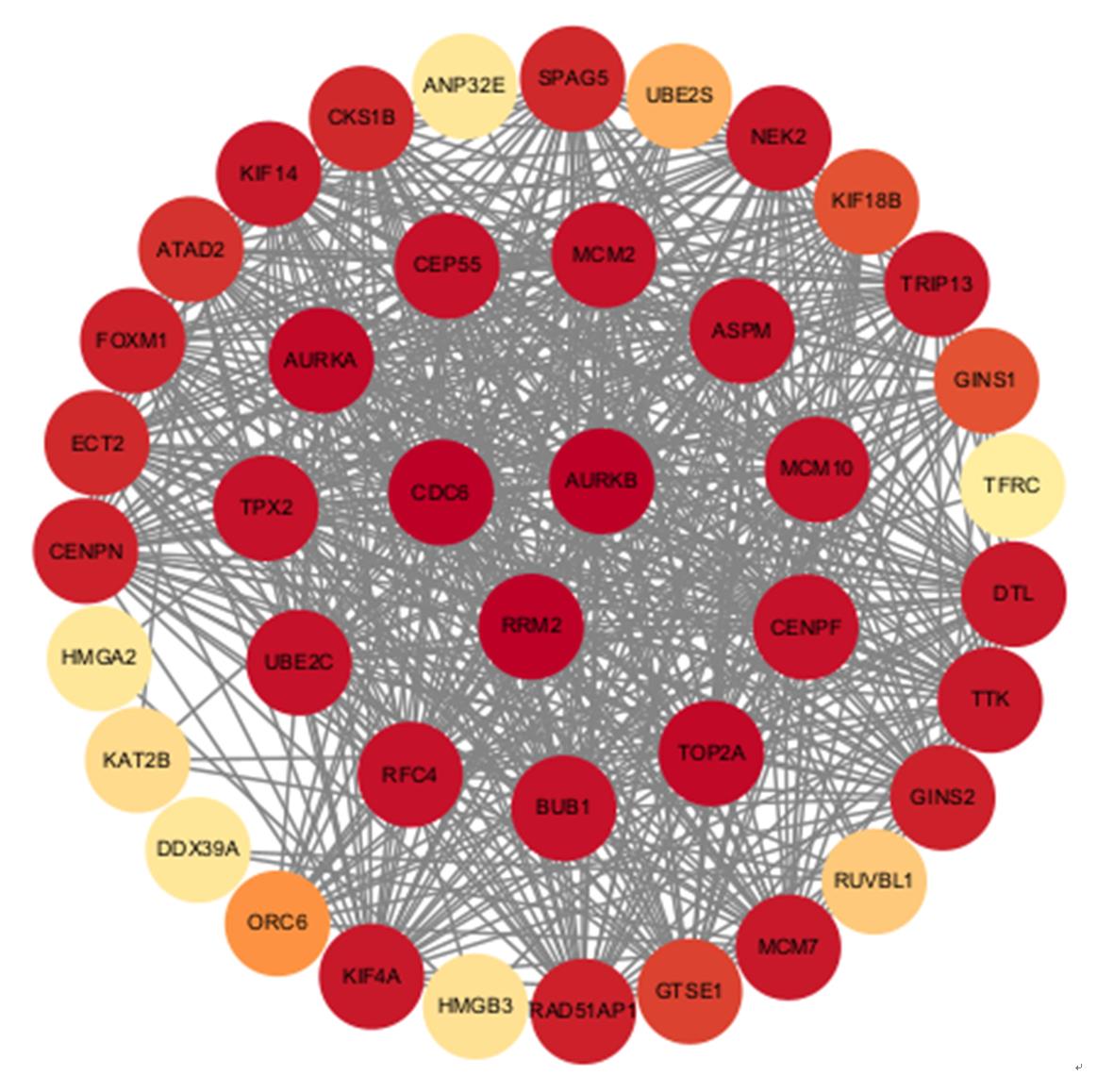
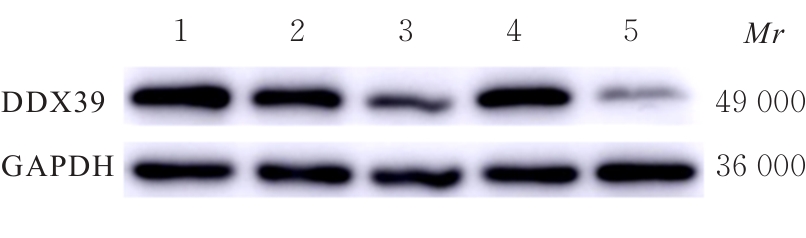
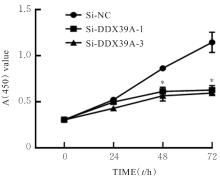
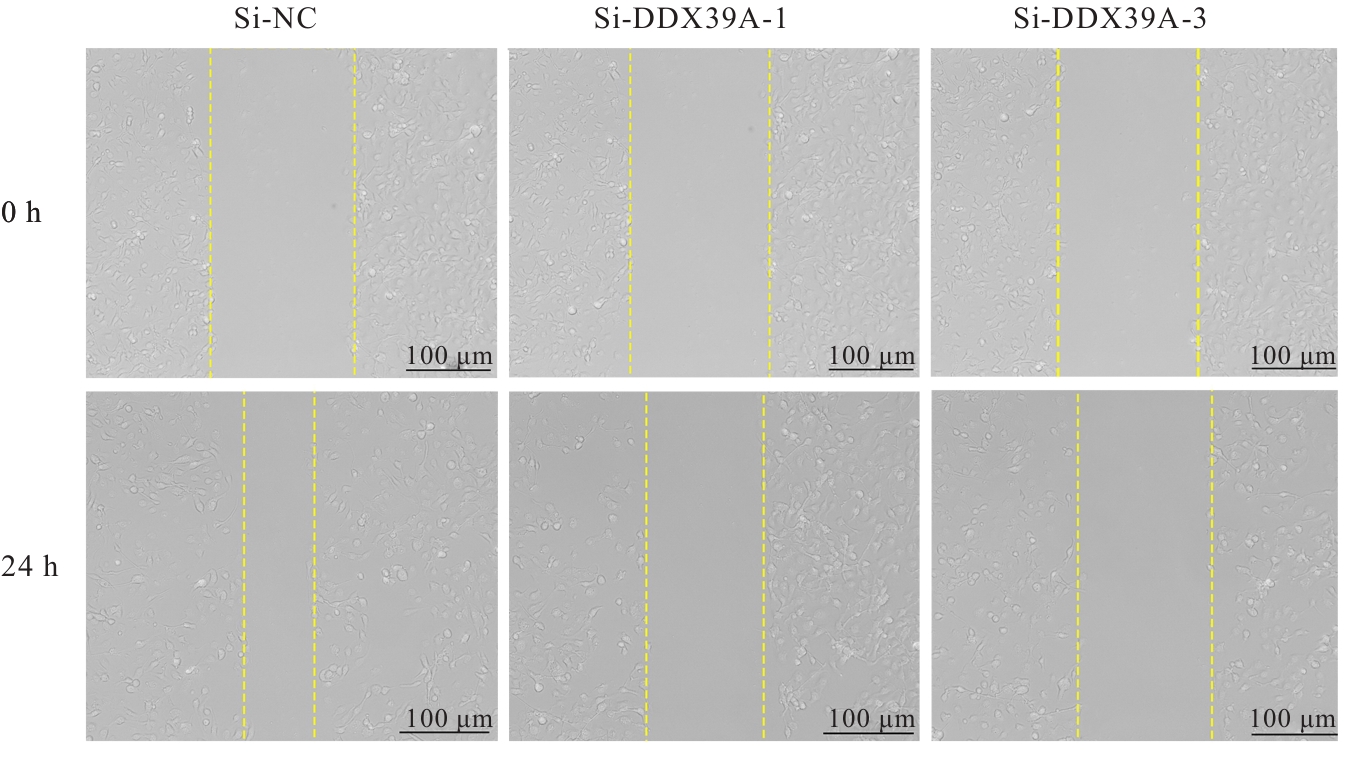
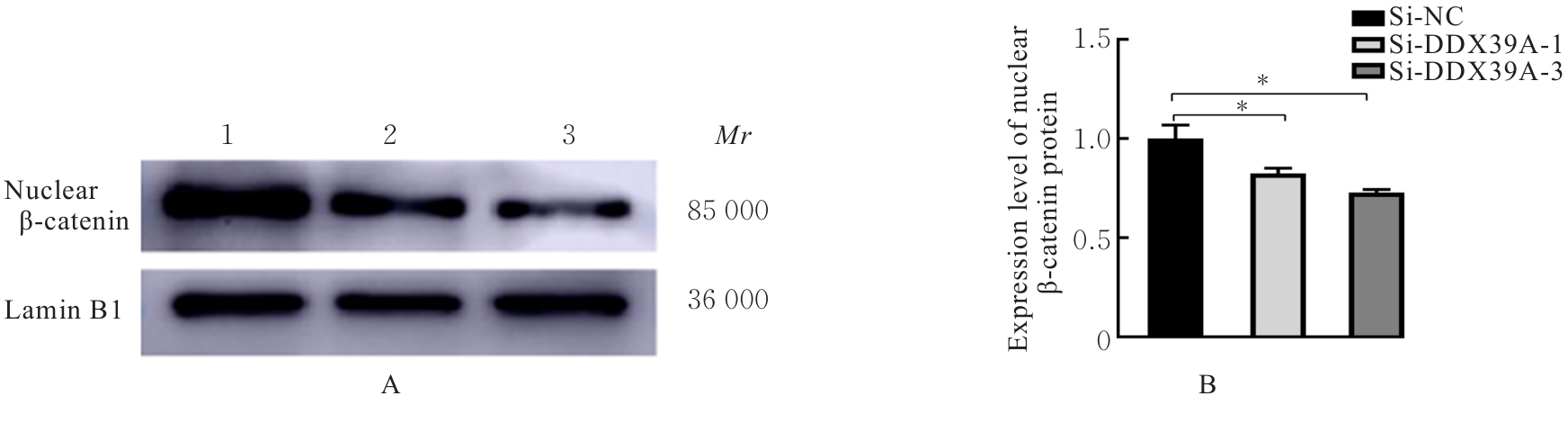
目的 探讨沉默DEAD-box RNA解旋酶39A(DDX39A)对食管癌TE-1细胞增殖、迁移和侵袭的调控作用,并阐明其可能的作用机制。 方法 通过基因表达汇编(GEO)数据库下载GSE63941、GSE77861、GSE20347和GSE16153芯片数据,利用癌症基因组图谱(TCGA)数据库筛选并下载食管癌相关数据;采用R软件分析差异表达基因;通过基因与蛋白质相互作用关系检索工具(STRING)构建蛋白-蛋白相互作用(PPI)网络数据,并采用Cytoscape软件中MCODE插件整合数据,鉴别出相关性强的关键基因,用基因表达谱数据动态分析网页工具(GEPIA 2)分析正常食管黏膜和食管癌组织中关键基因的表达情况,用卡普兰-迈尔曲线图(Kaplan-Meier Plotter)将筛查出来的关键基因进行生存分析并绘图。细胞学实验,选择食管癌TE-1细胞作为研究对象,采用小干扰RNA技术(siRNA)沉默DDX39A基因表达;取对数生长期TE-1细胞,将细胞分为空白组(MOCK组)、阴性对照组(si-NC组)和沉默组(si-DDX39A组);采用实时荧光定量PCR(RT-qPCR)和Western blotting法检测各组细胞中DDX39A mRNA及蛋白表达水平,CCK-8法检测各组细胞增殖活性,细胞划痕愈合实验检测各组细胞迁移率,Transwell小室实验检测各组TE-1细胞的侵袭细胞数,Western blotting法检测各组细胞中β-黏连蛋白(β-catenin)、糖原合成酶激酶-3β(GSK3β)、磷酸化糖原合成酶激酶-3β(p-GSK3β)、原癌基因(c-MYC)和细胞周期蛋白D1(Cyclin D1)及核内β-catenin蛋白表达水平。 结果 TCGA数据库与GEO数据库相结合共得到56个差异表达基因。Cytoscape软件MCODE插件共鉴别出41个相关性强的关键基因,通过GEPIA 2和Kaplan-Meier plotter数据库分析41个基因得到DDX39A。RT-qPCR和Western blotting法,与si-NC组比较,si-DDX39A组细胞中DDX39A mRNA和蛋白表达水平均降低(P<0.05)。CCK-8法,si-DDX39A组细胞增殖活性低于si-NC组(P<0.05);细胞划痕实验,24 h后,si-DDX39A组细胞迁移率低于si-NC组(P<0.05);Transwell小室实验,si-DDX39A组侵袭细胞数低于si-NC组(P<0.05)。与si-NC组比较,si-DDX39A-1组和si-DDX39A-3组TE-1细胞中β-catenin、p-GSK3β、c-MYC及Cyclin D1以及核内β-catenin蛋白表达水平均降低(P<0.01),而GSK3β蛋白表达水平差异无统计学意义(P>0.05)。 结论 DDX39A基因沉默可以抑制食管癌TE-1细胞的增殖、迁移和侵袭能力,其作用机制可能与调控Wnt/β-catenin信号通路有关。
中图分类号:
- R735.1