吉林大学学报(医学版) ›› 2024, Vol. 50 ›› Issue (5): 1313-1321.doi: 10.13481/j.1671-587X.20240515
• 基础研究 • 上一篇
基于酵母双杂交技术对与纳尔逊海湾正呼肠孤病毒σNS相互作用宿主蛋白的筛选
- 锦州医科大学基础医学院病原生物学教研室,辽宁 锦州 121000
Screening of host proteins interacting with Nelson Bay orthoreovirus σNS based on yeast two-hybrid technology
Lyuyin SUN,Zhuping MA,Runlin LI,Yonggang LI,Xiaoli TAO( )
)
- Department of Pathogenic Biology,School of Basic Medical Sciences,Jinzhou Medical University,Jinzhou 121000,China
摘要:
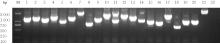
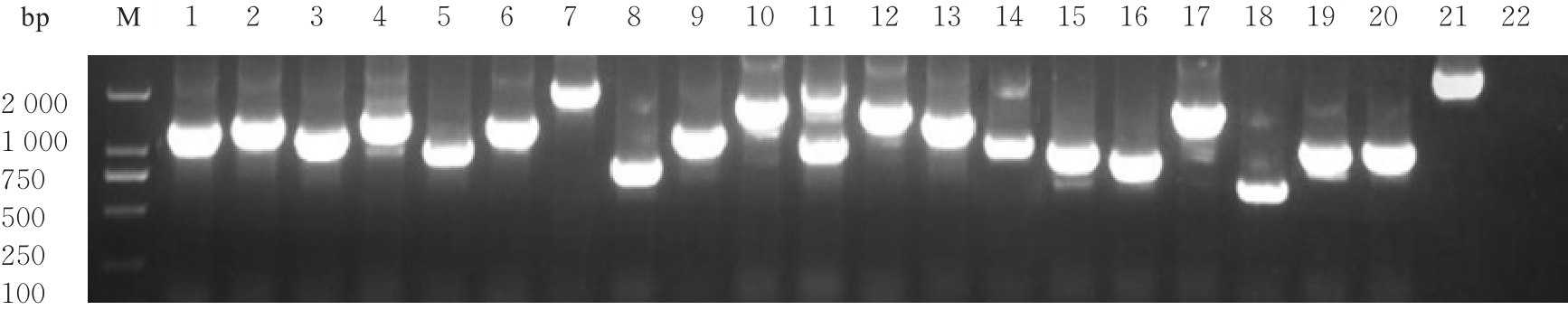
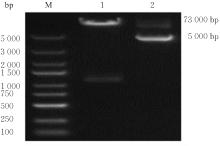
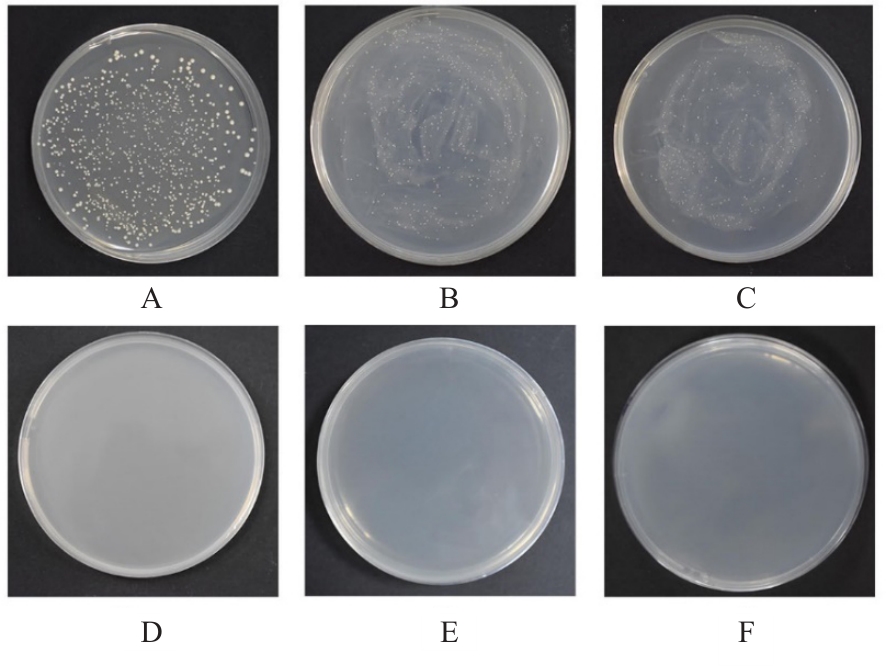
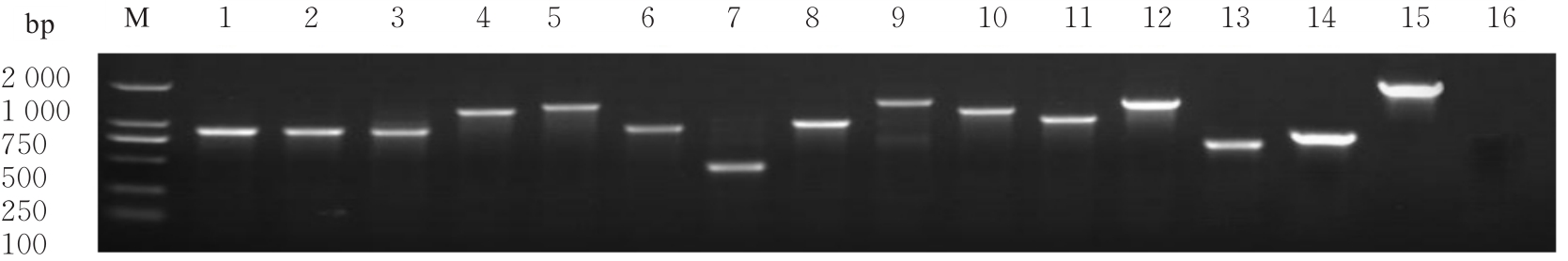
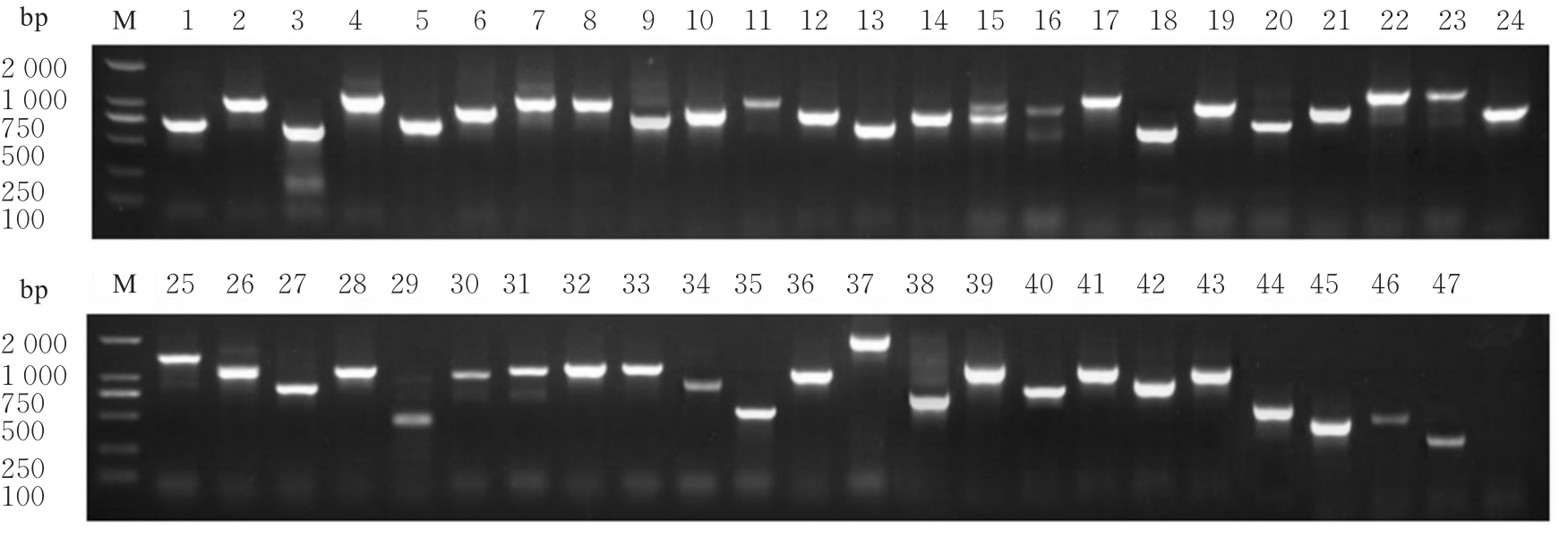
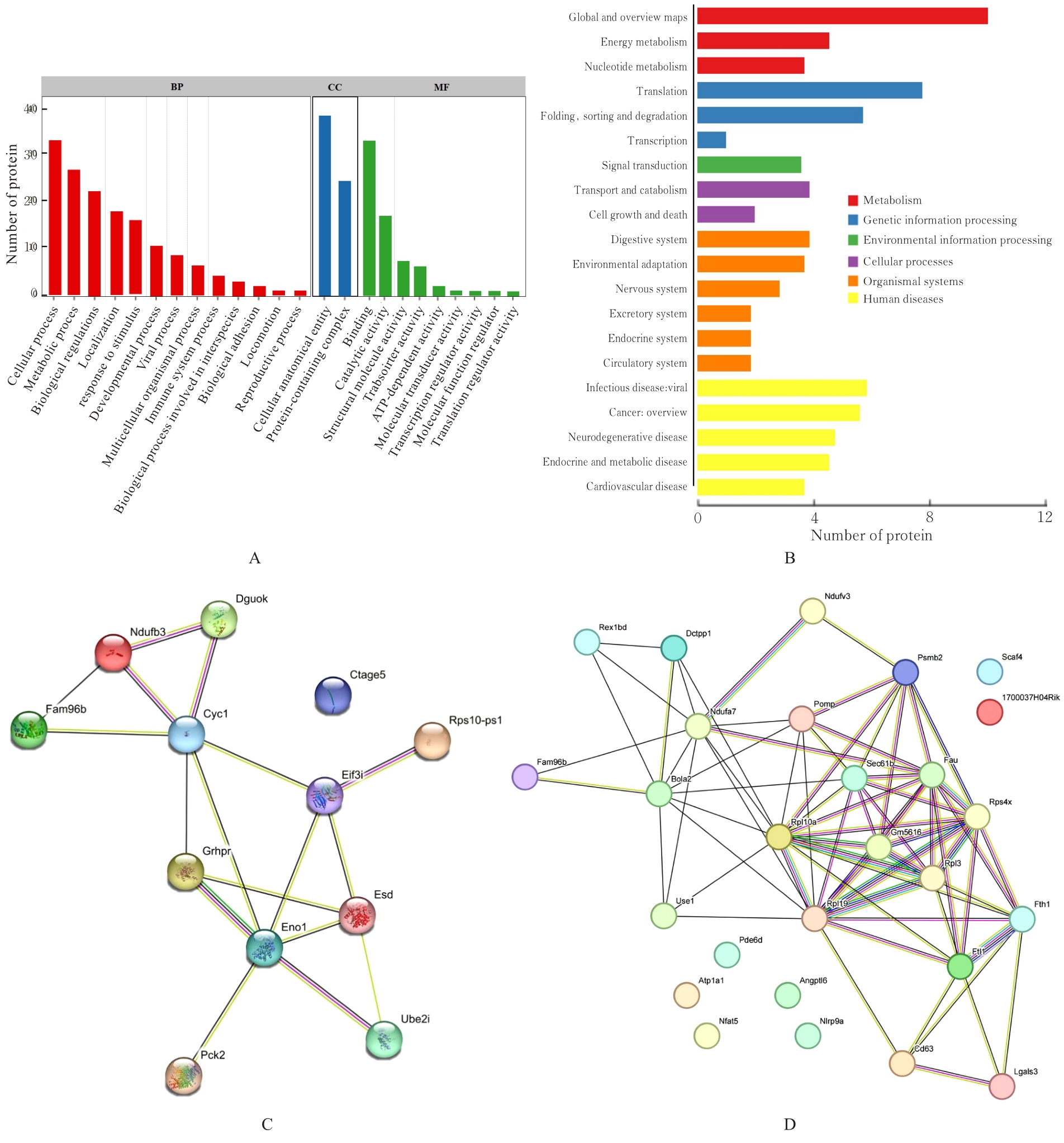
目的 探讨小鼠成纤维L929细胞中与纳尔逊海湾正呼肠孤病毒(NBV)σNS蛋白相互作用(互作)的宿主蛋白,阐明宿主蛋白对病毒复制的影响。 方法 构建表达σNS蛋白的诱饵质粒pGBKT7-S3,采用测序技术验证诱饵质粒在Y2H酵母细胞中的准确表达。将pGBKT7-S3和pGADT7质粒分别和共同转化至Y2HGold酵母细胞中,涂布于固体培养基进行培养,观察菌落生长情况,确认σNS蛋白对酵母细胞无毒性,不能自行激活报告基因。诱饵质粒pGBKT7-S3与小鼠成纤维L929细胞的cDNA文库进行杂交,对编码互作蛋白的阳性克隆进行质粒抽提,阳性测序结果通过Uniprot数据库检索筛选得到与NBV σNS互作的蛋白。对互作蛋白进行基因本体论(GO)功能富集分析、京都基因和基因组百科全书(KEGG)信号通路富集分析和STRING生物信息学分析。 结果 成功筛选出61个阳性克隆。DNA测序分析和BLAST比对分析去除23个未匹配到数据库和测序基本相同的阳性克隆。阳性测序结果通过Uniprot数据库检索筛选得到38个与NBV σNS互作的蛋白,31个蛋白参与细胞生物过程,36个蛋白是细胞解剖成分,31个蛋白具有结合功能,5个蛋白是线粒体呼吸链组成部分,7个蛋白是核糖体蛋白和核糖体大小亚基的组成部分,2个蛋白参与铁代谢稳态。GO功能富集分析,互作蛋白参与的生物过程(BP)富集在细胞过程、代谢过程、生物学调节、定位和对刺激的反应等;细胞组分主要是细胞解剖成分和含蛋白复合物;分子功能集中在结合、催化活性、结构分子活性和转运活性等方面。KEGG信号通路富集分析,蛋白在遗传信息处理途径的翻译、折叠、分类和降解信号通路中高度富集,在机体系统中主要与消化系统有关联;与多种病毒性传染病和癌症有关联。STRING分析,互作蛋白涉及核糖体蛋白类、蛋白修饰类、代谢类和免疫蛋白类等功能蛋白。 结论 成功筛选出与NBV σNS蛋白互作的蛋白,宿主蛋白在病毒复制中具有重要作用。
中图分类号:
- R373