Journal of Jilin University(Medicine Edition) ›› 2024, Vol. 50 ›› Issue (3): 739-748.doi: 10.13481/j.1671-587X.20240318
• Research in basic medicine • Previous Articles
Bioinformatics anlysis based on three-dimensional structure of Helicobacter pylori hp0169 gene
Linghui LIN1,Na LI1,Xiaoyan YIN1,Xiaoling WANG1,Yaping HU1,Wei LIU2,Rui FEI3( ),Xinli TIAN1(
),Xinli TIAN1( )
)
- 1.Department of Pathogenic Biology and Immunology,Xingtai Medical College,Xingtai 054000,China
2.Institute of Immunology,School of Basic Medical Scienes,Army Medical University,Chongqing 400038,China
3.Department of Cell Biology,School of Basic Medical Scienes,Jilin University,Changchun 130021,China
-
Received:2023-06-02Online:2024-05-28Published:2024-07-01 -
Contact:Rui FEI,Xinli TIAN E-mail:feirui@jlu.edu.cn;xttxl66@163.com
CLC Number:
- R378.99
Cite this article
Linghui LIN,Na LI,Xiaoyan YIN,Xiaoling WANG,Yaping HU,Wei LIU,Rui FEI,Xinli TIAN. Bioinformatics anlysis based on three-dimensional structure of Helicobacter pylori hp0169 gene[J].Journal of Jilin University(Medicine Edition), 2024, 50(3): 739-748.
share this article
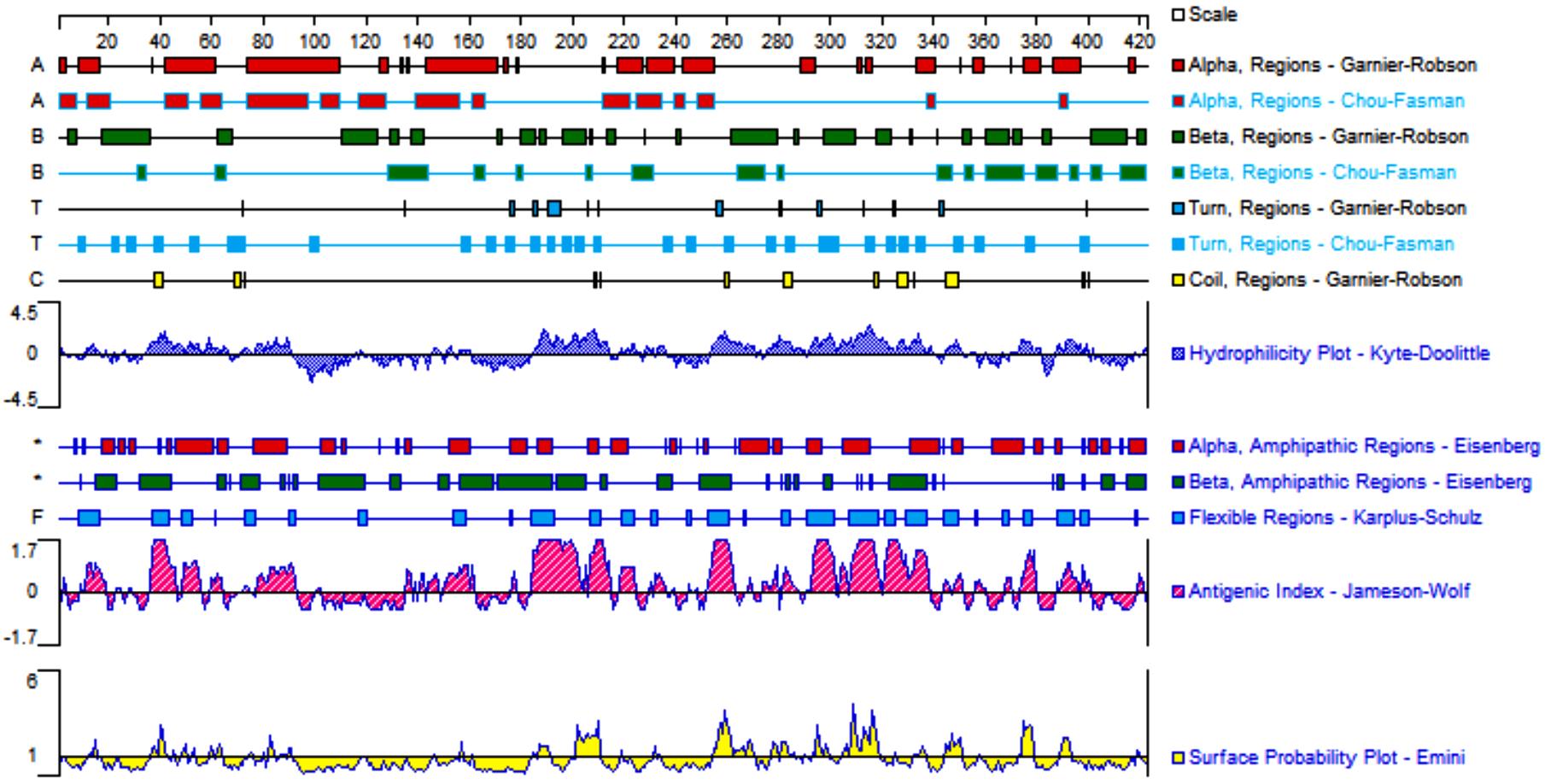
Tab.1
Antigenic epitopes of B lymphocytes of HpPrtC protein predicted by ABCpred software"
| Number | Amino acid sequence | Start postion | Score |
|---|---|---|---|
| 1 | GGVSHFSLRNRAGKEF | 29 | 0.89 |
| 2 | NGEITEDGRFFACKFT | 329 | 0.88 |
| 3 | KIEGRTKSSYYAAQTT | 252 | 0.88 |
| 4 | RGSCANDCRFDYEYYV | 191 | 0.88 |
| 5 | HIDKMAELEPDAFIIA | 82 | 0.87 |
| 6 | TTTTNTAYEIIAPKNA | 344 | 0.86 |
| 7 | QNHQTAISEGDFQVNG | 315 | 0.86 |
| 8 | YAAQTTRIYRLAVDDF | 262 | 0.86 |
| 9 | SGHIAEILSSNAISAL | 236 | 0.86 |
| 10 | KEGIDYAHALNKKVYA | 50 | 0.85 |
| 11 | AGKEFTLETFKEGIDY | 40 | 0.85 |
| 12 | LVEEEGVGTHIFNAKD | 217 | 0.85 |
Tab.2
Construct epitopes of HpPrtC protein predicted by IEDB software"
| Number | Residues | Number of residues | Score |
|---|---|---|---|
| 1 | A:R306, A:P308, A:F309 | 3 | 0.835 |
2 | A:F200, A:D201, A:Y202, A:E203, A:Y204, A:Y205, A:V206, A:K207, A:N208, A:P209, A:D210, A:N211, A:G212, A:V213, A:M214, A:M215, A:R216, A:L217, A:V218, A:E219, A:E220, A:E221, A:G222, A:V223, A:G224, A:T225, A:H226, A:N229, A:Q315, A:N316, A:H317, A:Q318, A:T319, A:A320 | 34 | 0.808 |
3 | A:T333, A:D335, A:G336, A:R337, A:F338, A:K342, A:F343, A:T344, A:T345, A:T346, A:T347, A:N348, A:I349, A:A350, A:Y351, A:K357, A:N358, A:A359, A:A360, A:I361, A:T362, A:P363, A:I364, A:V365, A:N366, A:E367, A:I368, A:G369, A:K370, A:I371, A:Y372, A:T373, A:S378, A:Y379, A:L380, A:V381, A:L382, A:Y383, A:K384, A:I385, A:L386, A:L387, A:E388, A:N389, A:N390, A:T391, A:E392, A:L393, A:E394, A:T395, A:I396, A:H397, A:S398, A:G399, A:N400, A:V401, A:N402, A:L403, A:V404, A:R405, A:L406, A:P407, A:A408, A:P409, A:L410, A:P411 | 66 | 0.742 |
| 4 | A:V420, A:E421, A:S422, A:K423, A:N424, A:G425, A:V426 | 7 | 0.738 |
5 | A:Q3, A:V31, A:S32, A:H33, A:F34, A:K50, A:D54, A:H57, A:G69, A:F70, A:P71, A:F72, A:N73, A:S74, A:Q75, A:L76, A:K77, A:L78, A:L79, A:E80, A:E81, A:H82, A:I83, A:Y84, A:K85, A:A87, A:E88, A:L89, A:E90, A:P91, A:A98, A:P99, A:G100, A:V101, A:V102, A:K103, A:L104, A:A105, A:L106, A:K107, A:I108, A:A109, A:P110, A:H111, A:I112, A:V122, A:L123, A:N124, A:L125, A:L126, A:D127, A:A128, A:Q129, A:V130, A:F131, A:Y132, A:D133, A:L134, A:G135, A:V136, A:K137, A:L145, A:S146, A:L147, A:N148, A:D149, A:A150, A:I151, A:E152, A:I153, A:K154, A:K155, A:A156, A:L157, A:P158, A:N159, A:L160, A:A240, A:E241, A:S244, A:S245, A:N246, A:A247, A:I248, A:F277, A:Y278, A:L304 | 87 | 0.663 |
| 6 | A:D276, A:H279, A:N280, A:T281, A:L282, A:K283, A:P284, A:S285 | 8 | 0.661 |
| 7 | A:R307, A:E310, A:D313 | 3 | 0.613 |
| 8 | A:C178, A:L179, A:I180, A:A182, A:L183, A:Q184, A:K185, A:G186, A:R187 | 9 | 0.596 |
| 9 | A:F44, A:T45, A:L46, A:E47 | 4 | 0.523 |
Tab.3
Antigenic epitopes of T lymphocytes of HpPrtC protein predicted by SYFPEITHI software"
| Start postion | Amino acid sequence | Length | Score |
|---|---|---|---|
| 25 | DAVYGGVSHFSLRNR | 15 | 28 |
| 52 | GIDYAHALNKKVYAT | 15 | 28 |
| 258 | KSSYYAAQTTRIYRL | 15 | 28 |
| 284 | PSFYASELNTLKNRG | 15 | 28 |
| 103 | KLALKIAPHIPIHLS | 15 | 26 |
| 159 | NLELEIFVHGSMCFA | 15 | 26 |
| 236 | SGHIAEILSSNAISA | 15 | 26 |
| 248 | ISALKIEGRTKSSYY | 15 | 26 |
| 271 | RLAVDDFYHNTLKPS | 15 | 26 |
| 390 | NTELETIHSGNVNLV | 15 | 26 |
| 234 | NLSGHIAEI | 8 | 28 |
| 122 | VLNLLDAQV | 8 | 25 |
| 58 | ALNKKVYAT | 8 | 24 |
| 156 | ALPNLELEI | 8 | 24 |
| 320 | AISEGDFQV | 8 | 24 |
| 379 | YLVLYKILL | 8 | 23 |
| 58 | ALNKKVYATI | 9 | 25 |
| 107 | KIAPHIPIHL | 9 | 23 |
| 125 | LLDAQVFYDL | 9 | 23 |
| 395 | TIHSGNVNLV | 9 | 23 |
| 1 | 张秋月, 李培杰, 董全江, 等. 胃癌相关幽门螺杆菌毒力和粘附基因进化分析[J]. 中国病原生物学杂志, 2021, 16(2): 161-166. |
| 2 | 覃艳春, 黄衍强, 陆 钢, 等. 幽门螺杆菌感染性慢性胃炎模型小鼠肠道各区域的菌群分布特征及其机制[J]. 吉林大学学报(医学版), 2023, 49(2): 289-297. |
| 3 | 中华医学会消化病学分会幽门螺杆菌学组. 第六次全国幽门螺杆菌感染处理共识报告(非根除治疗部分)[J]. 中华消化杂志, 2022, 42(5): 289-303. |
| 4 | FITZGERALD R, SMITH S M. An overview of helicobacter pylori infection[J]. Methods Mol Biol, 2021, 2283: 1-14. |
| 5 | DIACONU S, PREDESCU A, MOLDOVEANU A, et al. Helicobacter pylori infection: old and new[J]. J Med Life, 2017, 10(2): 112-117. |
| 6 | ZHAO H L, JI X F, CHEN X X, et al. Functional study of gene hp0169 in Helicobacter pylori pathogenesis[J]. Microb Pathog, 2017, 104: 225-231. |
| 7 | BERNEGGER S, JARZAB M, WESSLER S, et al. Proteolytic landscapes in gastric pathology and cancerogenesis[J]. Int J Mol Sci, 2022, 23(5): 2419. |
| 8 | 赵慧琳, 吴玉龙, 荣倩玉, 等. 幽门螺杆菌定植因子HpPrtC胶原蛋白酶的异源表达及其功能活性研究[J]. 中国病原生物学杂志, 2018, 13(9): 958-962. |
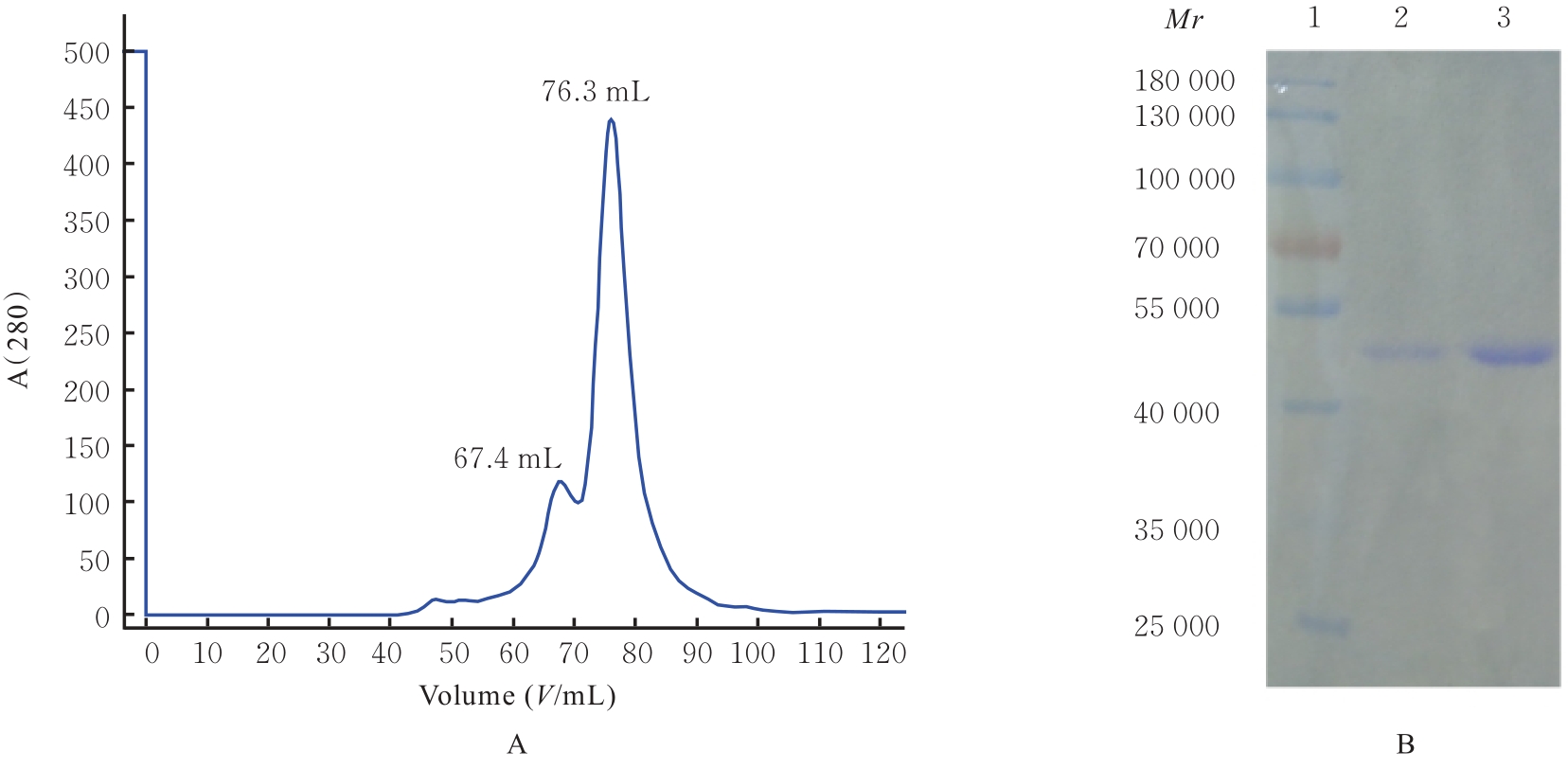
| 9 | 田新利, 刘 东, 郭 玲, 等. 幽门螺杆菌胶原蛋白酶HpPrtC的表达、纯化及结晶尝试[J]. 中国病原生物学杂志, 2018, 13(7): 719-723. |
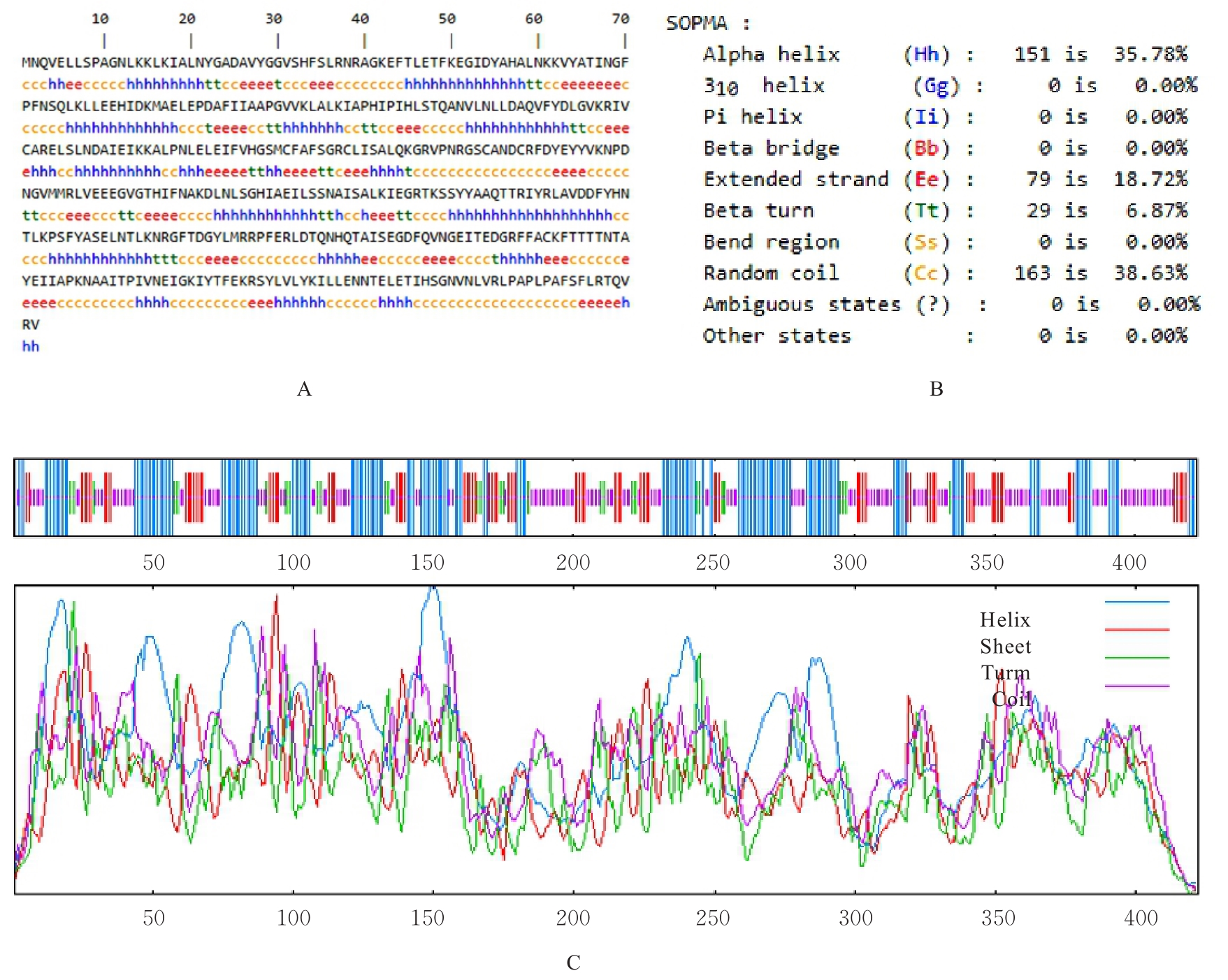
| 10 | GEOURJON C, DELÉAGE G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Bioinformatics, 1995, 11(6): 681-684. |
| 11 | THAWNG C N, SMITH G B. A transcriptome software comparison for the analyses of treatments expected to give subtle gene expression responses[J]. BMC Genomics, 2022, 23(1): 452. |
| 12 | 董 浩, 江海圳, 李 超, 等. 小鼠NR1D1基因过表达载体的构建及其生物信息学分析[J]. 吉林大学学报(医学版), 2022, 48(1): 94-103. |
| 13 | SCHACHERL M, MONTADA A A, BRUNSTEIN E, et al. The first crystal structure of the peptidase domain of the U32 peptidase family[J]. Acta Crystallogr D Biol Crystallogr, 2015, 71(Pt 12): 2505-2512. |
| 14 | MENDES M, MAHITA J, BLAZESKA N, et al. IEDB-3D 2.0: structural data analysis within the Immune Epitope Database[J]. Protein Sci, 2023, 32(4): e4605. |
| 15 | AHMAD MALIK A, OJHA S C, SCHADUANGRAT N, et al. ABCpred: a webserver for the discovery of acetyl-and butyryl-cholinesterase inhibitors[J]. Mol Divers, 2022, 26(1): 467-487. |
| 16 | SCHULER M M, NASTKE M D, STEVANOVIKĆ S. SYFPEITHI: database for searching and T-cell epitope prediction[J]. Methods Mol Biol, 2007, 409: 75-93. |
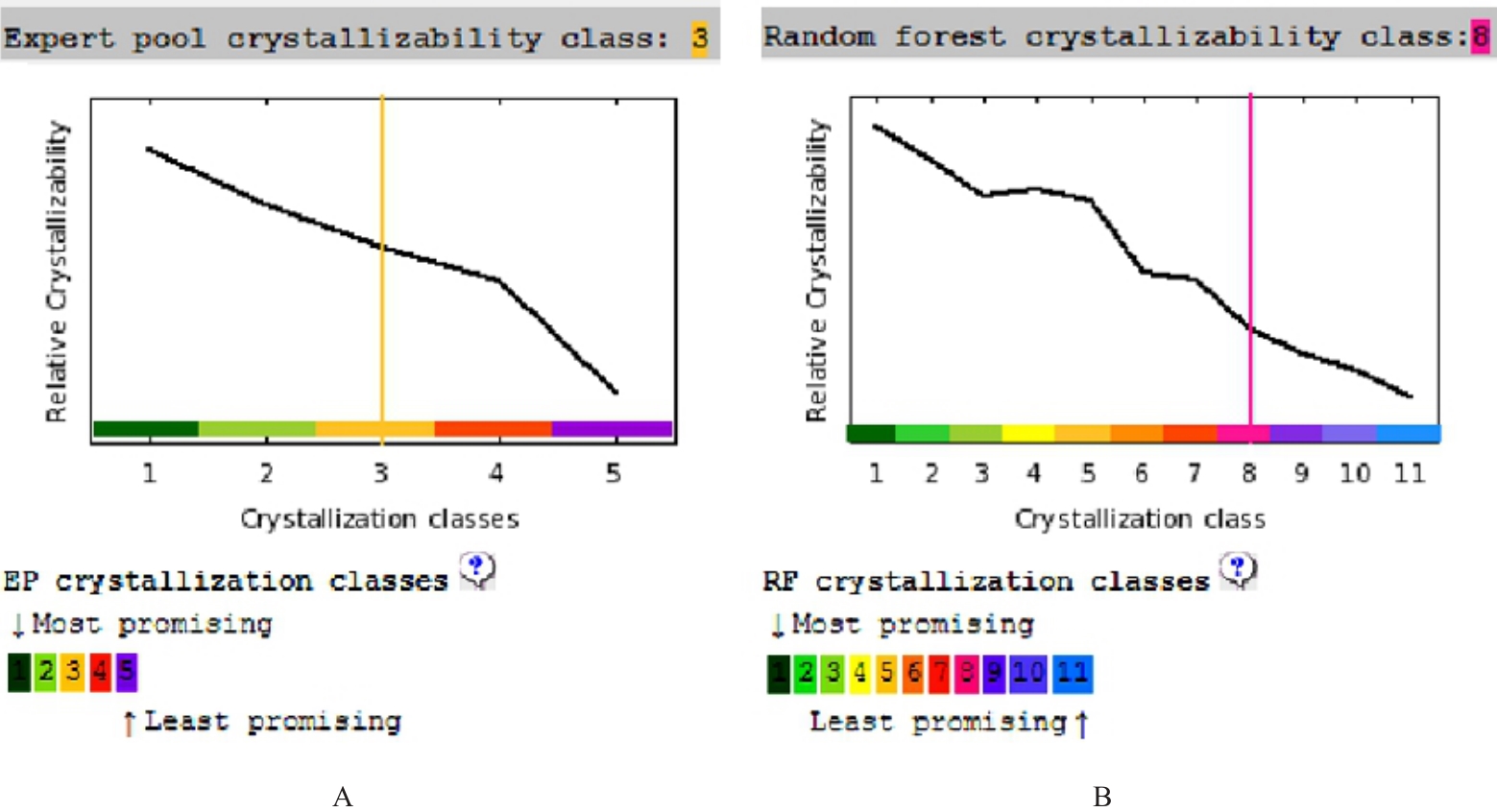
| 17 | SLABINSKI L, JAROSZEWSKI L, RYCHLEWSKI L, et al. XtalPred: a web server for prediction of protein crystallizability[J]. Bioinformatics, 2007, 23(24): 3403-3405. |
| 18 | ZHENG D D, LIANG S D, ZHANG C. B-cell epitope predictions using computational methods[J]. Methods Mol Biol, 2023, 2552: 239-254. |
| 19 | 金 昭, 王儒帅, 王朝阳, 等. 梅花草提取物对幽门螺杆菌小鼠胃黏膜定植的影响及其机制[J]. 吉林大学学报(医学版), 2022, 48(5): 1131-1138. |
| 20 | 常明珠, 李雨澎, 母润红, 等. 幽门螺旋杆菌检测方法及其应用价值的研究进展[J]. 吉林大学学报(医学版), 2023, 49(1): 253-260. |
| 21 | SAXENA A, MUKHOPADHYAY A K, NANDI S P. Helicobacter pylori: perturbation and restoration of gut microbiome[J]. J Biosci, 2020, 45(1): 110. |
| 22 | ZAGARI R M, FRAZZONI L, MARASCO G, et al. Treatment of Helicobacter pylori infection: a clinical practice update[J]. Minerva Med, 2021, 112(2): 281-287. |
| 23 | BOON L, UGARTE-BERZAL E, VANDOOREN J, et al. Protease propeptide structures, mechanisms of activation, and functions[J]. Crit Rev Biochem Mol Biol, 2020, 55(2): 111-165. |
| 24 | CROWCROFT N S, KLEIN N P. A framework for research on vaccine effectiveness[J]. Vaccine, 2018, 36(48): 7286-7293. |
| 25 | PALMER W H, NORMAN P J. The impact of HLA polymorphism on herpesvirus infection and disease[J]. Immunogenetics, 2023, 75(3): 231-247. |
| 26 | 韩 雪, 寸怡娜, 朱兰芳, 等. 甲型流感病毒T细胞表位预测及HLA-A2限制性抗原肽筛选[J]. 贵州医科大学学报, 2022, 47(5): 530-535, 541. |
| 27 | RAMANA J, MEHLA K. Immunoinformatics and epitope prediction[J]. Methods Mol Biol, 2020, 2131: 155-171. |
| 28 | 张舒阳, 毕研贞, 刘守胜, 等. 慢性乙型肝炎与HBV相关慢加急性肝衰竭患者血浆外泌体差异miRNA的生物信息学分析[J]. 临床肝胆病杂志, 2023, 39(8): 1848-1856. |
| 29 | MENDES M, MAHITA J, BLAZESKA N, et al. IEDB-3D 2.0: structural data analysis within the Immune Epitope Database[J]. Protein Sci, 2023, 32(4): e4605. |
| 30 | 柏晓辉, 刘 雪, 朱雯培, 等. 肺炎链球菌耐药相关蛋白Sp_0010生物信息学分析及结晶尝试[J]. 中国病原生物学杂志, 2020, 15(11): 1268-1271, 1276. |
| 31 | LIU H, ZHAO Y, SUN J. Heterogeneous nucleation in protein crystallization[J]. Biomimetics, 2023, 8(1): 68. |
| 32 | 郭 玲, 章金勇, 刘 东, 等. 幽门螺杆菌黏附素HpaA重组蛋白的表达、 纯化及晶体生长[J]. 免疫学杂志, 2015, 31(11): 986-990. |
| [1] | Yuting LIU,Ying YU,Guizhen LI,Qinxue SHI,Binbin LI. Bioinformatics analysis based on expression of splicing factor SRSF9 in head and neck squamous cell carcinoma and clinical significance [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 379-391. |
| [2] | Liping CHEN,Li HAN,Hua BIAN,Liye PANG. Bioinformatics analysis based on differentially expressed genes and screening of traditional Chinese medicine for treatment of severe bronchial asthma [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 411-421. |
| [3] | Minqi NING,Yong HE,Bingshu LI,Guotao HUANG,Xiaohu ZUO,Zhihan ZHAO,Wuyue HAN,Li HONG. Bioinformatics analysis based on pelvic organ prolapse related aging genes of GEO Database and LASSO regression algorithm [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 178-187. |
| [4] | Zixu YANG,Chang SU,Boyuan WANG,Chong LIU,Minghe LI. Bioinformatics analysis on molecular subtypes and clinical characteristics of head and neck squamous cell carcinoma based on genes associated with lactate metabolism [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 198-207. |
| [5] | Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252. |
| [6] | Yiming ZHAO,Haiyang XU. Bioinformatics analysis on related genes and candidate pathways of glioblastoma multiforme [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1280-1289. |
| [7] | Xiaoying ZHAO,Jiansheng SU,Jiahui MA,Yuefeng WANG,Dan WANG,Liyuan SUN. Bioinformatics analysis based on activity of heterotrimeric peptide H5LL and construction of recombinant Lactic acid bacteria expression vector [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1366-1374. |
| [8] | Yanchun QIN,Yanqiang HUANG,Gang LU,Ganrong HUANG,Huaying TANG,Yuanyuan DAI. Distribution characteristics of micrflora in various regions of intestinal tract of mice with chronic gastritis infected with Helicobacter Pylori andits mechannism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 289-297. |
| [9] | Xiaoyan WANG,Yihong HU,Yucheng HAN,Xianqiong ZOU. Bioinformatics analysis on mechanism of COMMD7 in occurrence and development of brain low-grade glioma [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 414-424. |
| [10] | Rui WANG,Ding ZHANG,Ruijian ZHUGE,Qian XUE,Li GUO. Bioinformatics analysis on mechanism of liver injury induced by hexavalent chromium [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 452-459. |
| [11] | Zhiyuan XIAO,Bing SONG,Xinyu MA,Lianhui JIN,Tong ZHENG,Fang CHAI. Bioinformatics analysis on expression of apoptosis related gene CD44 in thyroid carcinoma tissue and its relationship with tumor invasion and immune cell infiltration [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 473-481. |
| [12] | Wuyue TANG,Shuojie LI,Lijuan PANG. Bioinformatics analysis of splicing factor-alternative splicing regulatory network based on alternative splicing data in lung adenocarcinoma tissue [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 139-149. |
| [13] | Zhao JIN,Rushuai WANG,Zhaoyang WANG,zhanbiao HE,Fuming TIAN,Ruijun WANG. Effect of Parnassia palustris Linn extracts on Helicobacter pylori colonization in gastric mucosa and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1131-1138. |
| [14] | Xiaoyan WANG,Qiuyue ZHANG,Yujie HU,Zhijing MO. Bioinformatics analysis based on molecular characteristics and mechanism of cystic fibrosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1290-1297. |
| [15] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
|
||