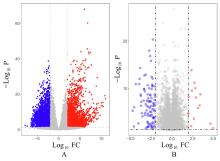
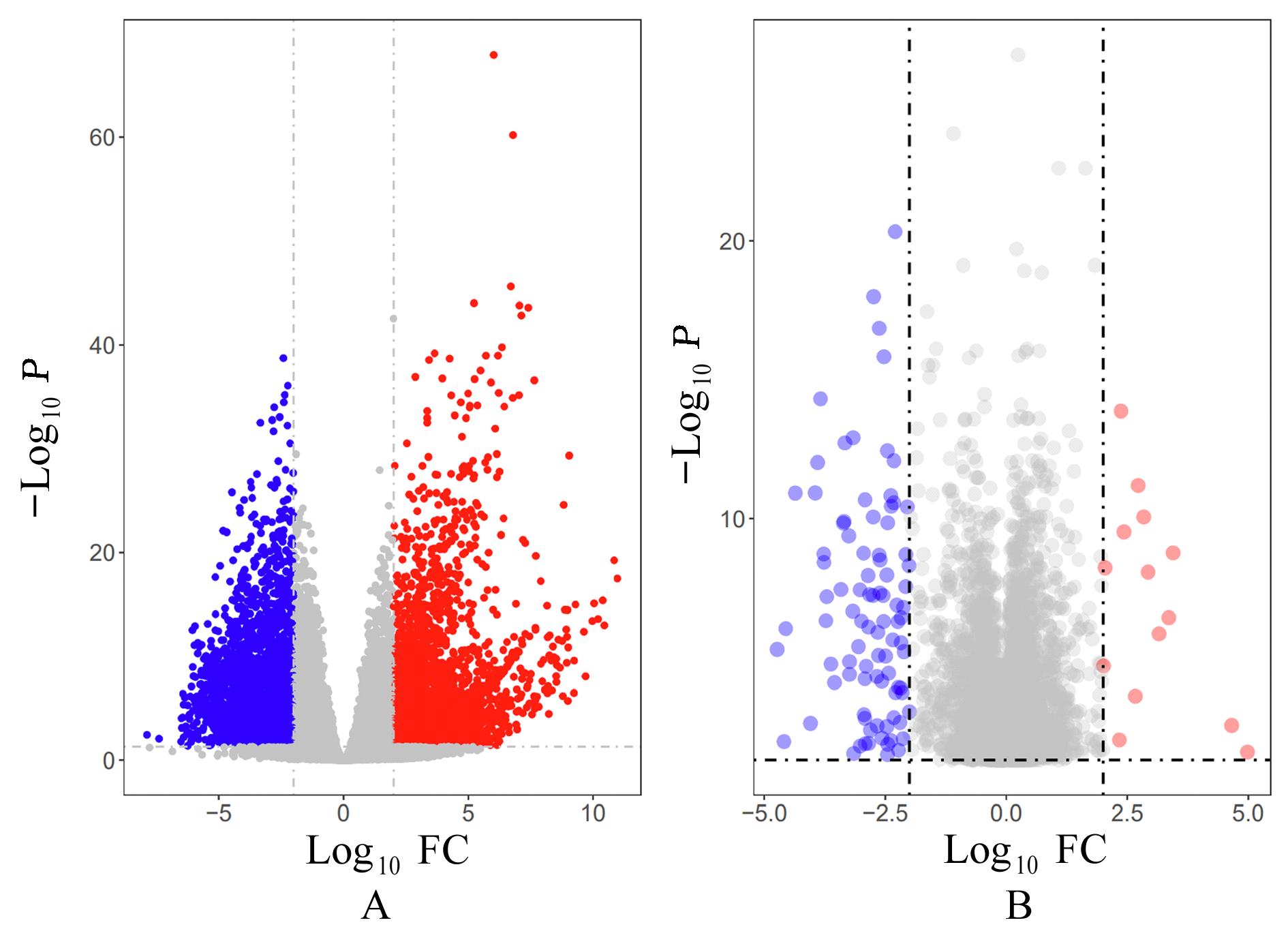
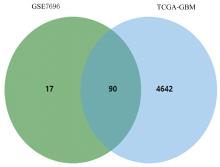
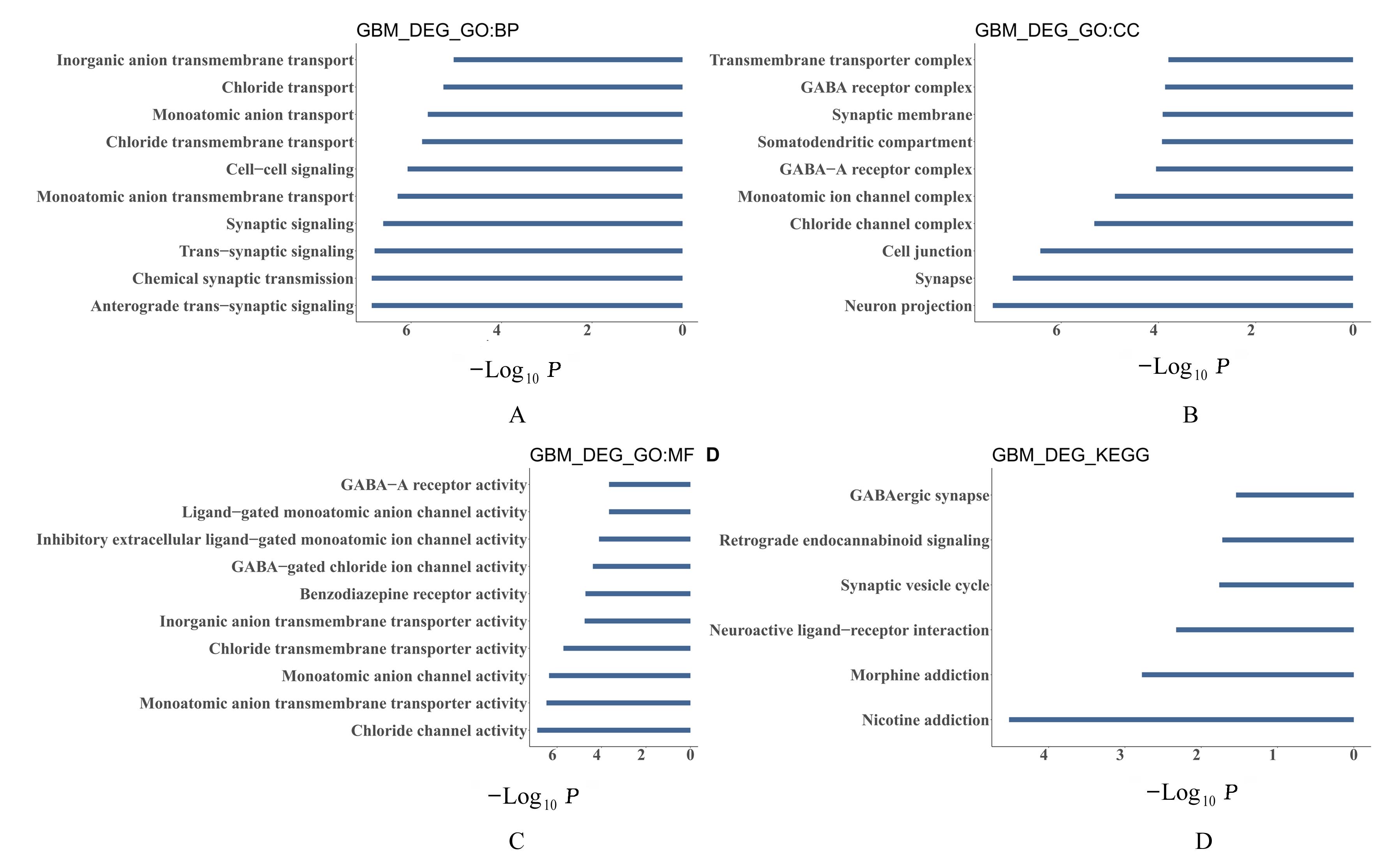
Journal of Jilin University(Medicine Edition) ›› 2023, Vol. 49 ›› Issue (5): 1280-1289.doi: 10.13481/j.1671-587X.20230522
• Research in clinical medicine • Previous Articles
Bioinformatics analysis on related genes and candidate pathways of glioblastoma multiforme
- Department of Neurosurgery,First Hospital,Jilin University,Changchun 130021,China
-
Received:2022-10-18Online:2023-09-28Published:2023-10-26 -
Contact:Haiyang XU E-mail:xuhaiyang76@163.com
CLC Number:
- R739.4
Cite this article
Yiming ZHAO,Haiyang XU. Bioinformatics analysis on related genes and candidate pathways of glioblastoma multiforme[J].Journal of Jilin University(Medicine Edition), 2023, 49(5): 1280-1289.
share this article
Tab.1
UDEGs in TCGA-GBM and GSE7696 Datasets"
| Gene | TCGA-GBM | GSE7696 | ||||||
|---|---|---|---|---|---|---|---|---|
| Log10 FC | P | Adjusted P | Log10 FC | P | Adjusted P | |||
| C7orf57 | 4.638 783 383 | 1.19E-06 | 8.13E-06 | 4.982 584 151 | 0.004 593 217 | 0.025 571 522 | ||
| CA3 | 6.086 673 206 | 1.29E-10 | 2.01E-09 | 2.336 620 217 | 0.001 249 899 | 0.009 301 135 | ||
| CENPK | 5.063 095 482 | 8.57E-38 | 7.26E-35 | 3.445 449 580 | 1.55E-11 | 1.72E-09 | ||
| CRNDE | 4.492 664 869 | 2.48E-19 | 1.61E-17 | 2.838 408 580 | 5.09E-13 | 8.77E-11 | ||
| DEPDC1B | 3.179 211 865 | 2.07E-12 | 4.57E-11 | 2.666 131 659 | 1.57E-05 | 0.000 249 488 | ||
| KIF23 | 4.169 152 072 | 3.31E-25 | 5.47E-23 | 3.158 584 562 | 3.68E-08 | 1.41E-06 | ||
| MEOX2 | 6.143 702 121 | 2.34E-16 | 9.55E-15 | 4.657 715 130 | 0.000 283 553 | 0.002 816 094 | ||
| NDC80 | 6.344 541 479 | 5.22E-44 | 1.68E-40 | 2.724 074 275 | 2.49E-14 | 6.50E-12 | ||
| NUF2 | 4.656 251 004 | 1.50E-30 | 5.55E-28 | 3.358 372 252 | 7.72E-09 | 3.70E-07 | ||
| RRM2 | 6.796 093 271 | 3.92E-65 | 6.31E-61 | 2.371 362 878 | 1.89E-17 | 1.36E-14 | ||
| TOP2A | 7.051 634 959 | 2.97E-48 | 1.59E-44 | 2.930 018 064 | 9.64E-11 | 8.48E-09 | ||
| TRIM5 | 2.864 864 227 | 1.68E-20 | 1.32E-18 | 2.040 196 151 | 6.45E-11 | 5.93E-09 | ||
| VIM | 3.649 502 968 | 2.41E-43 | 6.48E-40 | 2.434 029 893 | 2.16E-12 | 3.03E-10 | ||
Tab.1
Tab.2
DDEGs in TCGA-GBM and GSE7696 Datasets"
| Gene | TCGA-GBM | GSE7696 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log10 FC | P | Adjusted P | Log10 FC | P | Adjusted P | ||||||||
| ANO3 | -4.519 058 6 | 1.384 3E-11 | 2.624 1E-10 | -2.074 326 0 | 3.875E-10 | 2.807 5E-08 | |||||||
| ANXA3 | -2.444 998 2 | 5.048 3E-05 | 0.000 232 43 | -2.748 723 0 | 9.252 1E-10 | 5.885 8E-08 | |||||||
| ARPP21 | -3.735 909 4 | 2.871 1E-11 | 5.130 6E-10 | -3.053 561 9 | 1.247 4E-07 | 4.066 9E-06 | |||||||
| B3GALT2 | -2.124 367 2 | 0.000 224 14 | 0.000 881 05 | -2.448 459 8 | 0.001 990 35 | 0.013 462 15 | |||||||
| C11orf87 | -4.577 569 0 | 5.995 4E-10 | 8.111 7E-09 | -3.414 734 4 | 5.201 5E-10 | 3.570 2E-08 | |||||||
| C3orf80 | -3.739 123 2 | 2.530 1E-10 | 3.684 8E-09 | -3.726 593 3 | 1.050 9E-08 | 4.735 9E-07 | |||||||
| CACNA1G | -2.047 598 1 | 0.000 619 02 | 0.002 170 40 | -2.565 319 6 | 0.001 046 62 | 0.008 058 62 | |||||||
| CDH12 | -3.805 576 0 | 3.269 5E-05 | 0.000 157 11 | -2.172 632 9 | 7.758 6E-09 | 3.713 1E-07 | |||||||
| CLCA4 | -3.233 664 5 | 0.001 364 74 | 0.004 357 13 | -2.229 339 9 | 6.433 7E-06 | 0.000 118 93 | |||||||
| CLIC5 | -2.516 864 8 | 1.569 5E-06 | 1.038 8E-05 | -2.000 026 5 | 7.321 1E-05 | 0.000 910 38 | |||||||
| CNTN3 | -4.056 306 0 | 1.154 2E-05 | 6.199 3E-05 | -2.841 621 7 | 0.001 791 75 | 0.012 433 23 | |||||||
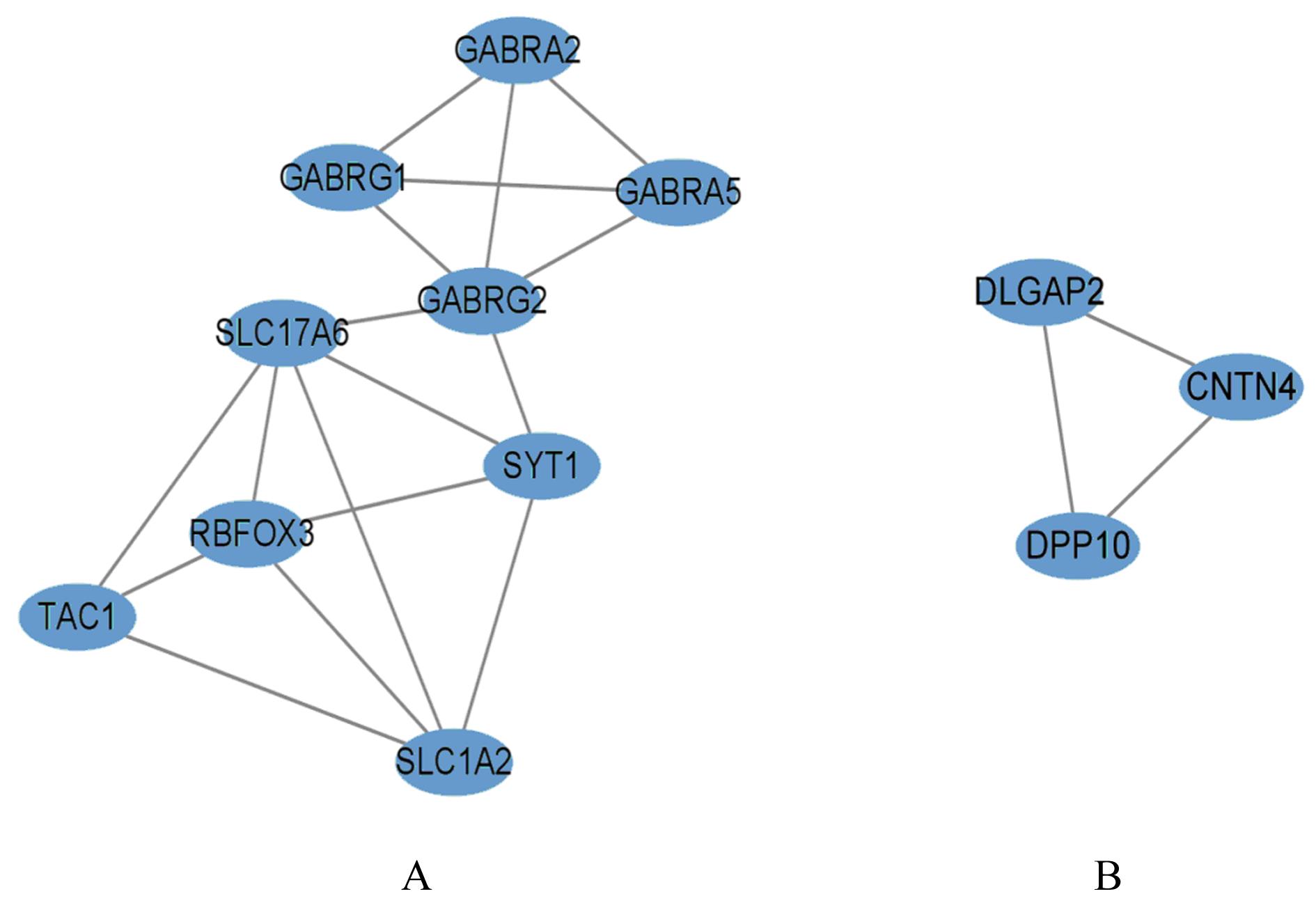
| CNTN4 | -3.921 157 7 | 1.578 7E-09 | 1.981 9E-08 | -2.843 849 1 | 1.945 9E-08 | 8.056E-07 | |||||||
| CNTNAP4 | -3.594 616 7 | 1.093 8E-06 | 7.506 7E-06 | -2.176 896 6 | 0.025 941 7 | 0.092 545 67 | |||||||
| CPEB3 | -4.146 164 0 | 6.467 7E-27 | 1.477 1E-24 | -3.171 898 4 | 4.283 7E-09 | 2.180 1E-07 | |||||||
| CYP4X1 | -4.013 594 9 | 6.120 9E-13 | 1.478 6E-11 | -2.947 741 1 | 1.605E-11 | 1.762 6E-09 | |||||||
| DACH2 | -4.267 847 7 | 4.436 6E-07 | 3.333 2E-06 | -3.239 297 2 | 1.839 1E-06 | 4.069 4E-05 | |||||||
| DCTN1-AS1 | -4.618 639 7 | 2.560 9E-09 | 3.070 1E-08 | -3.334 260 9 | 3.920 7E-16 | 1.900 6E-13 | |||||||
| DLGAP2 | -5.469 864 2 | 6.415 9E-11 | 1.061 7E-09 | -2.322 279 8 | 1.283 4E-13 | 2.674 7E-11 | |||||||
| DNAJA4 | -3.088 812 5 | 9.593 3E-17 | 4.220 1E-15 | -2.667 756 0 | 0.000 292 75 | 0.002 896 37 | |||||||
| DOK6 | -4.005 650 9 | 7.758 1E-10 | 1.022 6E-08 | -2.671 966 1 | 2.257 1E-06 | 4.840 3E-05 | |||||||
| DPP10 | -2.970 827 0 | 0.000 118 57 | 0.000 499 82 | -2.385 548 5 | 0.001 329 89 | 0.009 788 21 | |||||||
| EPHX4 | -2.665 576 7 | 1.199 8E-11 | 2.305 2E-10 | -2.008 055 6 | 5.333 9E-11 | 4.968 6E-09 | |||||||
| FAXC | -2.648 315 6 | 2.269 7E-13 | 5.922 8E-12 | -2.240 413 3 | 1.146 8E-08 | 5.113 2E-07 | |||||||
| FEZF2 | -4.097 037 3 | 1.658 2E-05 | 8.576 5E-05 | -2.544 726 5 | 9.380 7E-10 | 5.931 5E-08 | |||||||
| FSTL5 | -4.219 627 1 | 4.189 9E-07 | 3.163 4E-06 | -2.111 421 6 | 2.041 4E-07 | 6.196 1E-06 | |||||||
| FUT9 | -2.435 225 5 | 0.000 170 11 | 0.000 688 14 | -2.286 069 6 | 1.096 3E-05 | 0.000 185 07 | |||||||
| GABRA2 | -3.630 545 1 | 1.253 7E-06 | 8.482 6E-06 | -2.854 141 0 | 1.335 1E-10 | 1.127 9E-08 | |||||||
| GABRA5 | -5.557 260 5 | 1.186E-08 | 1.241 5E-07 | -3.770 595 9 | 1.747 6E-11 | 1.889 4E-09 | |||||||
| GABRG1 | -3.768 745 8 | 4.755 5E-05 | 0.000 220 24 | -2.322 539 5 | 0.000 130 02 | 0.001 474 64 | |||||||
| GABRG2 | -4.559 568 1 | 2.117 2E-06 | 1.359 5E-05 | -2.939 036 7 | 9.735 6E-05 | 0.001 153 57 | |||||||
| GALNTL5 | -4.328 213 1 | 0.000 893 32 | 0.002 997 68 | -3.166 021 0 | 2.417 1E-16 | 1.230 1E-13 | |||||||
| GJB6 | -4.259 117 6 | 0.000 371 55 | 0.001 382 35 | -3.239 197 5 | 5.164 3E-07 | 1.408 6E-05 | |||||||
| GNAL | -4.089 655 8 | 2.984E-13 | 7.626E-12 | -2.195 098 5 | 0.000 209 39 | 0.002 191 11 | |||||||
| GPR26 | -5.028 671 7 | 2.304 3E-05 | 0.000 115 22 | -2.746 869 4 | 5.188 1E-13 | 8.801 3E-11 | |||||||
| HCN1 | -5.010 596 0 | 9.773 5E-08 | 8.455 6E-07 | -4.556 066 5 | 2.259 9E-08 | 9.191 9E-07 | |||||||
| HTR2C | -5.977 880 4 | 2.088 4E-07 | 1.677 5E-06 | -2.917 951 7 | 9.938 4E-14 | 2.116 1E-11 | |||||||
| KLHL32 | -3.098 487 1 | 1.276 9E-07 | 1.076 6E-06 | -4.592 686 5 | 0.001 482 27 | 0.010 668 88 | |||||||
| KRT222 | -4.669 317 4 | 3.627E-05 | 0.000 172 57 | -2.623 954 8 | 8.874 5E-21 | 1.424 4E-17 | |||||||
| LINC00320 | -2.612 491 0 | 0.00 017 963 | 0.000 723 11 | -2.173 423 8 | 8.649 8E-08 | 2.954E-06 | |||||||
| LINC00507 | -5.248 378 4 | 2.066 8E-05 | 0.000 104 52 | -3.765 626 5 | 3.932 2E-11 | 3.798 6E-09 | |||||||
| LINC00622 | -2.262 619 3 | 1.943 8E-05 | 9.907 2E-05 | -2.182 898 9 | 6.969E-06 | 0.000 126 89 | |||||||
| MAL2 | -5.613 979 9 | 6.142 2E-08 | 5.554 2E-07 | -3.363 748 9 | 9.264 4E-13 | 1.475 6E-10 | |||||||
| MCF2L2 | -3.801 827 9 | 2.243 4E-11 | 4.086 3E-10 | -3.546 277 5 | 4.028 3E-06 | 8.051 1E-05 | |||||||
| NECAB1 | -3.121 465 6 | 1.335 1E-13 | 3.597 7E-12 | -3.254 491 2 | 3.131E-12 | 4.188E-10 | |||||||
| NPY1R | -3.412 642 3 | 7.229 5E-06 | 4.086 3E-05 | -2.921 630 2 | 2.817 5E-06 | 5.873 1E-05 | |||||||
| NWD2 | -6.093 546 7 | 7.362 2E-09 | 8.025 4E-08 | -2.739 299 5 | 5.404 8E-22 | 1.025 2E-18 | |||||||
| OLFM3 | -4.384 256 7 | 1.818E-07 | 1.483 5E-06 | -2.626 311 5 | 1.972 8E-11 | 2.089 5E-09 | |||||||
| OR2L13 | -3.184 174 3 | 0.00 137 556 | 0.004 385 57 | -2.432 026 6 | 1.62E-06 | 3.682 3E-05 | |||||||
| PAK3 | -3.891 071 8 | 3.527 6E-10 | 5.008 5E-09 | -2.803 053 9 | 0.000 454 05 | 0.004 137 21 | |||||||
| PAK5 | -3.262 887 5 | 0.000 481 18 | 0.001 736 86 | -4.044 749 9 | 0.000 229 26 | 0.002 362 32 | |||||||
| PCLO | -4.427 415 2 | 4.894 1E-10 | 6.758E-09 | -2.894 964 1 | 8.309 6E-07 | 2.071 5E-05 | |||||||
| PCSK2 | -3.449 592 2 | 0.000 150 43 | 0.000 616 36 | -2.533 642 6 | 1.129 5E-08 | 5.046 9E-07 | |||||||
| PDE1A | -3.786 849 9 | 1.798 9E-12 | 4.025 4E-11 | -2.262 699 7 | 2.305 1E-09 | 1.3E-07 | |||||||
| PTPN20 | -2.650 374 9 | 0.002 899 93 | 0.008 429 39 | -4.734 737 7 | 1.643 2E-07 | 5.132 8E-06 | |||||||
| PTPRR | -4.256 576 7 | 1.138 6E-09 | 1.459 5E-08 | -3.021 135 8 | 5.399 5E-10 | 3.669 9E-08 | |||||||
| RBFOX3 | -5.164 526 9 | 2.240 4E-09 | 2.725 5E-08 | -2.377 946 5 | 1.802 5E-13 | 3.582 1E-11 | |||||||
| RBM11 | -3.036 017 9 | 8.848 1E-05 | 0.000 383 99 | -2.386 307 1 | 6.901 1E-14 | 1.515 8E-11 | |||||||
| RBP4 | -4.133 621 7 | 2.541E-09 | 3.047 5E-08 | -2.041 955 9 | 1.942 2E-13 | 3.787 5E-11 | |||||||
| RGS7BP | -4.745 285 1 | 5.245 2E-17 | 2.378 9E-15 | -2.345 543 2 | 6.622 9E-08 | 2.346 2E-06 | |||||||
| RSPO2 | -3.054 422 4 | 0.000 214 95 | 0.000 848 75 | -2.821 181 4 | 8.497 2E-10 | 5.489 3E-08 | |||||||
| SERTM1 | -5.217 704 1 | 1.345 5E-07 | 1.129 5E-06 | -3.351 972 9 | 7.918 4E-13 | 1.280 8E-10 | |||||||
| SLC17A6 | -3.758 383 8 | 0.000 875 85 | 0.002 946 43 | -2.149 296 7 | 1.107 3E-05 | 0.000 186 49 | |||||||
| SLC1A2 | -3.567 993 4 | 4.151 1E-11 | 7.132 8E-10 | -2.469 439 2 | 0.000 319 60 | 0.003 097 40 | |||||||
| SLC2A13 | -2.823 002 3 | 1.141 6E-12 | 2.642 8E-11 | -2.455 376 9 | 0.006 058 37 | 0.031 611 38 | |||||||
| SLC35F3 | -4.564 621 5 | 1.908 7E-09 | 2.346 8E-08 | -2.451 721 1 | 8.888 7E-13 | 1.426 7E-10 | |||||||
| SLITRK4 | -3.895 418 5 | 4.602 7E-06 | 2.725 5E-05 | -2.612 023 4 | 7.267 4E-10 | 4.783 6E-08 | |||||||
| SPOCK3 | -3.500 679 4 | 5.818 5E-06 | 3.370 4E-05 | -2.639 120 8 | 2.938E-07 | 8.526 3E-06 | |||||||
| STYK1 | -5.752 268 9 | 4.477 2E-09 | 5.107E-08 | -3.836 853 7 | 6.106 4E-18 | 4.900 6E-15 | |||||||
| SYT1 | -5.297 412 2 | 7.312 9E-11 | 1.193 5E-09 | -3.708 976 9 | 1.044 9E-09 | 6.508 4E-08 | |||||||
| TAC1 | -2.413 136 2 | 0.019 040 18 | 0.042 920 05 | -3.154 706 4 | 0.005 396 08 | 0.028 922 32 | |||||||
| THEMIS | -3.642 492 0 | 3.349 3E-10 | 4.770 1E-09 | -2.295 036 8 | 1.118 7E-24 | 4.668 7E-21 | |||||||
| TRHDE | -5.301 796 0 | 1.860 6E-08 | 1.863 5E-07 | -3.899 271 2 | 2.802 4E-15 | 9.586 2E-13 | |||||||
| TSPYL1 | -2.808 882 4 | 3.369 1E-35 | 2.086 3E-32 | -2.071 533 8 | 1.833E-11 | 1.958 5E-09 | |||||||
| TYRP1 | -3.680 997 7 | 0.000 226 27 | 0.000 888 45 | -2.125 646 7 | 2.911 6E-09 | 1.594 6E-07 | |||||||
| UNC13C | -4.937 175 3 | 2.764 6E-08 | 2.676 6E-07 | -4.358 387 7 | 5.344 3E-14 | 1.212 1E-11 | |||||||
| VIP | -4.090 428 4 | 6.264 8E-08 | 5.657E-07 | -3.948 695 7 | 5.21E-14 | 1.194 6E-11 | |||||||
| WIF1 | -4.376 320 5 | 0.000 404 | 0.001 488 99 | -2.651 992 2 | 3.184 5E-08 | 1.246 7E-06 | |||||||
Tab.2
| 1 | GOODENBERGER M L, JENKINS R B. Genetics of adult glioma[J]. Cancer Genet, 2012, 205(12): 613-621. |
| 2 | LOUIS D N, PERRY A, REIFENBERGER G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary[J]. Acta Neuropathol, 2016, 131(6): 803-820. |
| 3 | MØLLER H G, RASMUSSEN A P, ANDERSEN H H,et al. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion[J].Mol Neurobiol, 2013, 47(1): 131-144. |
| 4 | VERHAAK R G W, HOADLEY K A, PURDOM E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA,IDH1,EGFR,and NF1[J]. Cancer Cell, 2010, 17(1): 98-110. |
| 5 | CLAUS E B, WALSH K M, WIENCKE J K, et al. Survival and low-grade glioma: the emergence of genetic information[J]. Neurosurg Focus, 2015, 38(1): E6. |
| 6 | ALIFIERIS C, TRAFALIS D T. Glioblastoma multiforme: pathogenesis and treatment[J]. Pharmacol Ther, 2015, 152: 63-82. |
| 7 | THAKKAR J P, DOLECEK T A, HORBINSKI C, et al. Epidemiologic and molecular prognostic review of glioblastoma[J]. Cancer Epidemiol Biomarkers Prev, 2014, 23(10): 1985-1996. |
| 8 | MAHATA B, BISWAS S, RAYMAN P, et al. GBM derived gangliosides induce T cell apoptosis through activation of the caspase cascade involving both the extrinsic and the intrinsic pathway[J]. PLoS One, 2015, 10(7): e0134425. |
| 9 | MA H W, LI T, TAO Z N, et al. NKCC1 promotes EMT-like process in GBM via RhoA and Rac1 signaling pathways[J]. J Cell Physiol, 2019, 234(2): 1630-1642. |
| 10 | CHEN X J, LIN Q X, JIANG Y A, et al. Identification of potential biomarkers of platelet RNA in glioblastoma by bioinformatics analysis[J]. Biomed Res Int, 2022, 2022: 2488139. |
| 11 | REN Z J, HE Y, YANG Q Q, et al. A comprehensive analysis of the glutathione peroxidase 8 (GPX8) in human cancer[J]. Front Oncol, 2022, 12: 812811. |
| 12 | YUAN F, CAI X M, CONG Z X, et al. Roles of the m6A modification of RNA in the glioblastoma microenvironment as revealed by single-cell analyses[J]. Front Immunol, 2022, 13: 798583. |
| 13 | LIU J H, GAO L, JI B W, et al. BCL7A as a novel prognostic biomarker for glioma patients[J]. J Transl Med, 2021, 19(1): 335. |
| 14 | YU T L, ACHARYA A, MATTHEOS N, et al. Molecular mechanisms linking peri-implantitis and type 2 diabetes mellitus revealed by transcriptomic analysis[J]. PeerJ, 2019, 7: e7124. |
| 15 | PAOLACCI S, PRECONE V, ACQUAVIVA F,et al. Genetics of lipedema: new perspectives on genetic research and molecular diagnoses[J]. Eur Rev Med Pharmacol Sci, 2019, 23(13): 5581-5594. |
| 16 | OMURO A, DEANGELIS L M. Glioblastoma and other malignant gliomas: a clinical review[J]. JAMA, 2013, 310(17): 1842-1850. |
| 17 | SIEGHART W. Structure and pharmacology of gamma-aminobutyric acid A receptor subtypes[J]. Pharmacol Rev, 1995, 47(2): 181-234. |
| 18 | BLANCHART A, FERNANDO R, HÄRING M,et al. Endogenous GABAA receptor activity suppresses glioma growth[J]. Oncogene, 2017, 36(6): 777-786. |
| 19 | BARBIERI F, BOSIO A G, PATTAROZZI A, et al. Chloride intracellular channel 1 activity is not required for glioblastoma development but its inhibition dictates glioma stem cell responsivity to novel biguanide derivatives[J]. J Exp Clin Cancer Res, 2022,41(1): 53. |
| 20 | OLSEN R W, SIEGHART W. International Union of Pharmacology. LXX. Subtypes of gamma-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function Update[J]. Pharmacol Rev, 2008, 60(3): 243-260. |
| 21 | MAILLARD P Y, BAER S, SCHAEFER É, et al. Molecular and clinical descriptions of patients with GABAA receptor gene variants (GABRA1, GABRB2, GABRB3, GABRG2): a cohort study, review of literature, and genotype-phenotype correlation[J]. Epilepsia, 2022, 63(10): 2519-2533. |
| 22 | ZHANG X M, JIANG M Y, TANG S F, et al. Comprehensive bioinformatic analysis of key genes and signaling pathways in glioma[J].JUSTC, 2022, 52(9): 3. |
| 23 | SÜDHOF T C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle[J]. Neuron, 2013, 80(3): 675-690. |
| 24 | NORD H, HARTMANN C, ANDERSSON R, et al. Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array[J]. Neuro-oncology, 2009, 11(6): 803-818. |
| 25 | MISAWA K, KANAZAWA T, MISAWA Y, et al. Frequent promoter hypermethylation of tachykinin-1 and tachykinin receptor type 1 is a potential biomarker for head and neck cancer[J]. J Cancer Res Clin Oncol, 2013, 139(5): 879-889. |
| 26 | WRANGLE J, MACHIDA E O, DANILOVA L,et al. Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer[J]. Clin Cancer Res, 2014, 20(7): 1856-1864. |
| 27 | DUAN W, ZHANG Y P, HOU Z, et al. Novel insights into NeuN: from neuronal marker to splicing regulator[J]. Mol Neurobiol, 2016, 53(3): 1637-1647. |
| 28 | LIU T Z, LI W B, LU W J, et al. RBFOX3 promotes tumor growth and progression via hTERT signaling and predicts a poor prognosis in hepatocellular carcinoma[J]. Theranostics, 2017, 7(12): 3138-3154. |
| 29 | MARAGAKIS N J, ROTHSTEIN J D. Glutamate transporters: animal models to neurologic disease[J]. Neurobiol Dis, 2004, 15(3): 461-473. |
| 30 | CONSORTIUM E. De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies[J]. Am J Hum Genet, 2016, 99(2): 287-298. |
| 31 | YAO P S, KANG D Z, LIN R Y, et al. Glutamate/glutamine metabolism coupling between astrocytes and glioma cells: neuroprotection and inhibition of glioma growth[J]. Biochem Biophys Res Commun, 2014, 450(1): 295-299. |
| [1] | Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252. |
| [2] | Rui WANG,Ding ZHANG,Ruijian ZHUGE,Qian XUE,Li GUO. Bioinformatics analysis on mechanism of liver injury induced by hexavalent chromium [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 452-459. |
| [3] | Xiaoyan WANG,Yihong HU,Yucheng HAN,Xianqiong ZOU. Bioinformatics analysis on mechanism of COMMD7 in occurrence and development of brain low-grade glioma [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 414-424. |
| [4] | Zhiyuan XIAO,Bing SONG,Xinyu MA,Lianhui JIN,Tong ZHENG,Fang CHAI. Bioinformatics analysis on expression of apoptosis related gene CD44 in thyroid carcinoma tissue and its relationship with tumor invasion and immune cell infiltration [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 473-481. |
| [5] | Wuyue TANG,Shuojie LI,Lijuan PANG. Bioinformatics analysis of splicing factor-alternative splicing regulatory network based on alternative splicing data in lung adenocarcinoma tissue [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 139-149. |
| [6] | Xiaoyan WANG,Qiuyue ZHANG,Yujie HU,Zhijing MO. Bioinformatics analysis based on molecular characteristics and mechanism of cystic fibrosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1290-1297. |
| [7] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
| [8] | Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765. |
| [9] | Xiaoyan LI,Wei ZHANG,Jie HE. Promotion effect of REG1A on proliferation and migration of lung adenocarcinoma cells by regulating Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 444-453. |
| [10] | Nan LI,Lei CHEN,Tianmin XU,Kun ZHANG. Bioinformatics analysis on screening of extracellular matrix related genes in patients with endometriosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 188-194. |
| [11] | Hao DONG,Haizhen JIANG,Chao LI,Dengke GAO,Yaping JIN,Huatao CHEN. Construction of mouse NR1D1 gene overexpression vector and its bioinformatics analysis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 94-103. |
| [12] | Zhijuan ZHAO,Lian MENG,Chunxia LIU. Bioinformatics analysis on miRNA-mRNA regulatory networks based on fusion genes acting in rhabdomyosarcoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 154-162. |
| [13] | Ying ZHAO,Danyu ZHAO,Chao LIU. Bioinformatics analysis on prognostic evaluation value of TXNDC11 gene in pan-cancer and its immunity regulation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 142-153. |
| [14] | Yang YU,Sainan LIU,Yunkai LIU,Yong LI,Yichun QIAO,Yi CHENG. Bioinformatic analysis on expression characteristics of PRPS2 and its relationship with prognosis of breast cancer [J]. Journal of Jilin University(Medicine Edition), 2021, 47(5): 1229-1236. |
| [15] | Yuqing PAN, Yunyan SUN, Yixun LI, Liying WANG, Zhuoma SINAN, Rui CHEN, Xi ZHANG, Yan DU. Construction and analysis of competitive endogenous RNA networks in acute myeloid leukemia based on high-throughput microarray [J]. Journal of Jilin University(Medicine Edition), 2021, 47(3): 669-676. |
|
||