Journal of Jilin University(Medicine Edition) ›› 2025, Vol. 51 ›› Issue (3): 703-715.doi: 10.13481/j.1671-587X.20250315
• Research in clinical medicine • Previous Articles
Bioinformatics analysis on adjustment effect of colorectal liver metastases model in mice based on complement alternative pathway and its experimental verification
Changyu SHI1,Yong LI1,Jing DENG1,Chunmei PIAO2,Ming JIN1( )
)
- 1.Department of Biochemistry and Molecular Biology,School of Medical Sciences,Yanbian University,Yanji 133000,China
2.Affiliated Beijing Anzhen Hospital,Capital Medical University,Beijing Institute of Heart Lung and Blood Vessel Diseases,Beijing 100029,China
-
Received:2024-05-09Accepted:2024-08-06Online:2025-05-28Published:2025-07-18 -
Contact:Ming JIN E-mail:jinming@ybu.edu.cn
CLC Number:
- R735.3
Cite this article
Changyu SHI,Yong LI,Jing DENG,Chunmei PIAO,Ming JIN. Bioinformatics analysis on adjustment effect of colorectal liver metastases model in mice based on complement alternative pathway and its experimental verification[J].Journal of Jilin University(Medicine Edition), 2025, 51(3): 703-715.
share this article
Tab.1
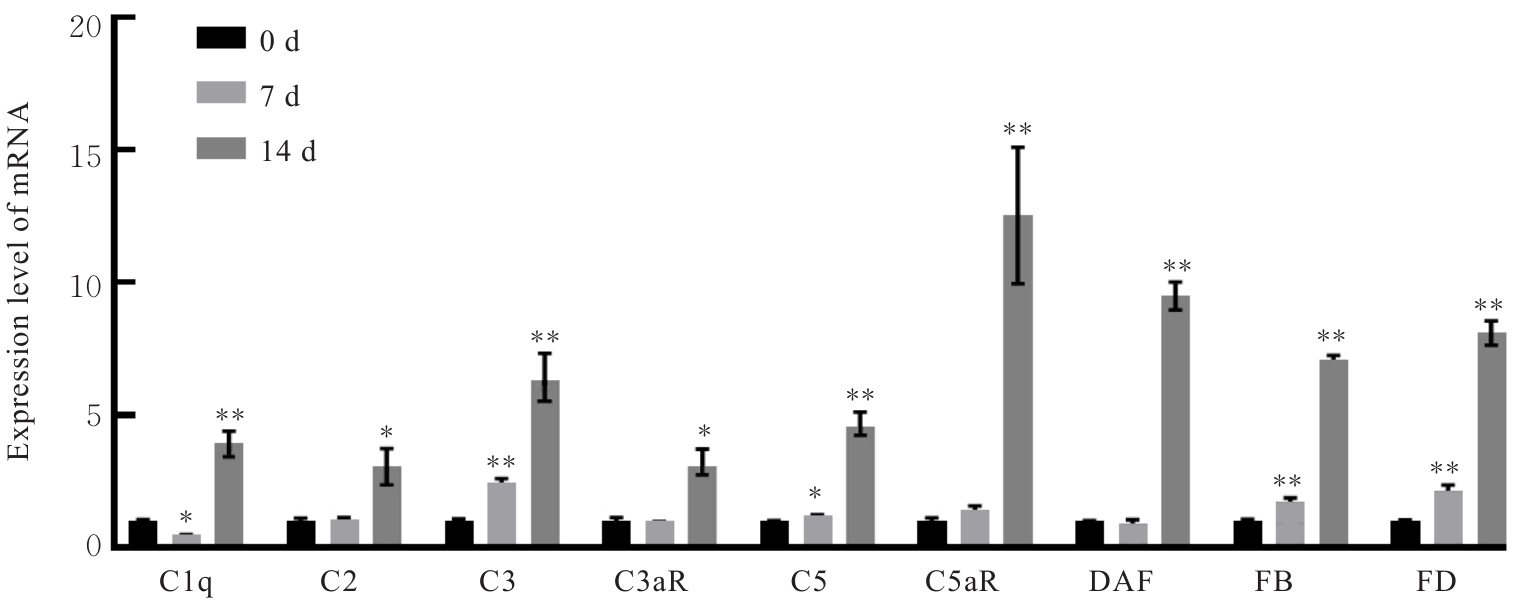
Sequences of primers"
| Primer | Forward(5'-3') | Reverse(5'-3') |
|---|---|---|
| C1q | GAAGGGCGTGAAAGGCAATC | CAAGCGTCATTGGGTTCTGC |
| C2 | AGAGTGCCGAACTCATGGTG | GGCTGAGAGGCAAAGGTGAT |
| C3 | ATAAAGAGCCAGCGGCTACA | GAATGAAGGGGTAAGGGGCA |
| C3aR | CCCCAAGACATTGCCTCCAT | GACTGTGTTCACGGTCGTCT |
| C5 | TGGTTCCTTCAGCACGACTC | CAGACTGCGTCAGCCACTAA |
| C5aR | TCCTTCAGAAGAGTTGCCTGC | TTCTGTGGTAACCAGCGACG |
| DAF | GCTCAATTAACTGCGGCTCAA | ACAGAGACAGCGACAGCAAC |
| FB | ATGGGGACAAGAAAGCCAGTT | TAGCCTTGGGCCTTTGTAGC |
| FD | TCGAAGGTGTGGTTACGTGG | TCGATCCACATCCGGTAGGA |
| GADPH | AATGCATCCTGCACCACC | ATGCCAGTGAGCTTCCCG |
| [1] | BENSON A B, VENOOK A P, AL-HAWARY M M, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology[J]. J Natl Compr Canc Netw, 2021, 19(3): 329-359. |
| [2] | TSAI M S, SU Y H, HO M C, et al. Clinicopathological features and prognosis in resectable synchronous and metachronous colorectal liver metastasis[J]. Ann Surg Oncol, 2007, 14(2): 786-794. |
| [3] | MAURI G, BONAZZINA E, AMATU A, et al. The evolutionary landscape of treatment for BRAFV600E mutant metastatic colorectal cancer[J]. Cancers (Basel), 2021, 13(1): 137. |
| [4] | RUFFINELLI J C, SANTOS VIVAS C, SANZ-PAMPLONA R, et al. New advances in the clinical management of RAS and BRAF mutant colorectal cancer patients[J]. Expert Rev Gastroenterol Hepatol, 2021, 15(1): 65-79. |
| [5] | LUO Q X, CHEN D K, FAN X J, et al. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: a single-site report[J]. Transl Oncol, 2020, 13(12): 100874. |
| [6] | WANG D W, SU F, YANG L J, et al. Bioinformatics analysis and identification of potential genes associated with pathogenesis and prognosis of gastric cancer[J]. Curr Med Sci, 2022, 42(2): 357-372. |
| [7] | TALAAT I M, ELEMAM N M, SABER-AYAD M. Complement system: an immunotherapy target in colorectal cancer[J]. Front Immunol, 2022, 13: 810993. |
| [8] | CHANG H L, JIN L, XIE P Y, et al. Complement C5 is a novel biomarker for liver metastasis of colorectal cancer[J]. J Gastrointest Oncol, 2022, 13(5): 2351-2365. |
| [9] | PIAO C M, CAI L, QIU S L, et al. Complement 5a enhances hepatic metastases of colon cancer via monocyte chemoattractant protein-1-mediated inflammatory cell infiltration[J]. J Biol Chem, 2015, 290(17): 10667-10676. |
| [10] | IRIZARRY R A, HOBBS B, COLLIN F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data[J]. Biostatistics, 2003, 4(2): 249-264. |
| [11] | WEI R Q, ZHANG W M, LIANG Z, et al. Identification of signal pathways and hub genes of pulmonary arterial hypertension by bioinformatic analysis[J]. Can Respir J, 2022, 2022: 1394088. |
| [12] | RITCHIE M E, PHIPSON B, WU D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Res, 2015, 43(7): e47. |
| [13] | YOUNG A, WHITEHOUSE N, CHO J, et al. OntologyTraverser: an R package for GO analysis[J]. Bioinformatics, 2005, 21(2): 275-276. |
| [14] | KANEHISA M. The KEGG database[J]. Novartis Found Symp, 2002, 247: 91-101; discussion101-103, 119-128, 244-252. |
| [15] | 崔 巍, 杜 杰, 杨 敏, 等. 小鼠结直肠癌肝转移模型的建立及其炎症微环境分析[J]. 中国癌症杂志, 2012, 22(8): 566-572, 582. |
| [16] | SCHILLING J D, MACHKOVECH H M, KIM A H J, et al. Macrophages modulate cardiac function in lipotoxic cardiomyopathy[J]. Am J Physiol Heart Circ Physiol, 2012, 303(11): H1366-H1373. |
| [17] | HAN Y H, MUN J G, JEON H D, et al. Betulin inhibits lung metastasis by inducing cell cycle arrest, autophagy, and apoptosis of metastatic colorectal cancer cells[J]. Nutrients, 2019, 12(1): 66. |
| [18] | MAREI H E, ALTHANI A, AFIFI N, et al. p53 signaling in cancer progression and therapy[J]. Cancer Cell Int, 2021, 21(1): 703. |
| [19] | ADMASU F T, DEJENIE T A, AYEHU G W, et al. Evaluation of thromboembolic event, basic coagulation parameters, and associated factors in patients with colorectal cancer: a multicenter study[J]. Front Oncol, 2023, 13: 1143122. |
| [20] | KROONE C, TIEKEN C, KOCATÜRK B, et al. Tumor-expressed factor Ⅶ is associated with survival and regulates tumor progression in breast cancer[J]. Blood Adv, 2023, 7(11): 2388-2400. |
| [21] | DAHA M R, SEELEN M. Novel approaches to control of the alternative complement pathway for the treatment of C3 glomerulopathies[J]. J Am Soc Nephrol, 2018, 29(8): 2032-2033. |
| [22] | COSS S L, ZHOU D L, CHUA G T, et al. The complement system and human autoimmune diseases[J]. J Autoimmun, 2023, 137: 102979. |
| [23] | AYKUT B, PUSHALKAR S, CHEN R N, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL[J]. Nature, 2019, 574(7777): 264-267. |
| [24] | 郭楠楠, 王兴智. 补体B因子的研究进展[J]. 国际免疫学杂志, 2022, 45(1): 6. |
| [25] | 路 平, 魏少忠, 梁新军. 补体系统与肿瘤免疫的研究进展[J]. 中国肿瘤临床, 2021, 48(13): 681-685. |
| [26] | 陈立颖, 朱 莹, 封 奕, 等. 结肠癌组织中补体C5a/C5aR通路活化上调FGL2的表达[J]. 第三军医大学学报, 2016, 38(6): 575-579. |
| [27] | WU P, SHI J Y, SUN W, et al. The prognostic value of plasma complement factor B (CFB) in thyroid carcinoma[J]. Bioengineered, 2021, 12(2): 12854-12866. |
| [28] | MANTOVANI A, ALLAVENA P, MARCHESI F, et al. Macrophages as tools and targets in cancer therapy[J]. Nat Rev Drug Discov, 2022, 21(11): 799-820. |
| [1] | Wenchang CAI,Yuqi LIU,Han WANG,Helin WANG,Zhenjiang WANG,Zishen XIAO,Shiyuan MA,Liping AN,Yanbo LIU. Expression of protein kinase D2 in bladder cancer tissue and its effect on tumor immune microenvironment [J]. Journal of Jilin University(Medicine Edition), 2025, 51(2): 378-391. |
| [2] | Bo LIU,Chao SUN,Xu WANG,Kewei MA. Bioinformatics analysis on differentially expressed genes in multiple primary lung cancers based on GEO database [J]. Journal of Jilin University(Medicine Edition), 2025, 51(2): 437-446. |
| [3] | Xiaoyan WANG,Xuelian LI,Bin LIANG,Wenfei TIAN,Hailin MA,Zhijing MO. Analysis on relationship between CALU and prognosis of hepatocellular carcinoma patients and its mechanism based on transcriptome and single cell sequencing data [J]. Journal of Jilin University(Medicine Edition), 2025, 51(2): 447-459. |
| [4] | Huaxia MU,Weixiao BU,Shuting DING,Mengyao GAO,Weiqiang SU,Zhen ZHANG,Qifu BO,Feng LIU,Fuyan SHI,Qinghua WANG,Yujia KONG,Suzhen WANG. Two sample Mendelian randomization study on causal relationship between insulin-like growth factor-1 and colorectal cancer [J]. Journal of Jilin University(Medicine Edition), 2025, 51(2): 479-485. |
| [5] | Pengli WU,Fengyu LI,Bo LIU,Yang LYU. Effect of silencing DDX39A gene on proliferation, migration and invasion of esophageal cancer TE-1 cells and its mechanism [J]. Journal of Jilin University(Medicine Edition), 2025, 51(1): 115-123. |
| [6] | Lyuyin SUN,Zhuping MA,Runlin LI,Yonggang LI,Xiaoli TAO. Screening of host proteins interacting with Nelson Bay orthoreovirus σNS based on yeast two-hybrid technology [J]. Journal of Jilin University(Medicine Edition), 2024, 50(5): 1313-1321. |
| [7] | Jinlian LI,Lanzhen HUANG,Xishi HUANG,Kangzhi LI,Jiali JIANG,Miaomiao ZHANG,Qunying WU. Bioinformatics analysis on key genes related to prognosis, diagnosis, and immune cell infiltration of hepatocellular carcinoma and their potential therapeutic drugs [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 1062-1075. |
| [8] | Yuanguo WANG,Peng ZHANG. Bioinformatics analysis based on relationship between SSP1 and TGFB1 and occurrence, prognosis, and immune invasion of esophageal adenocarcinoma [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 1076-1086. |
| [9] | Linghui LIN,Na LI,Xiaoyan YIN,Xiaoling WANG,Yaping HU,Wei LIU,Rui FEI,Xinli TIAN. Bioinformatics anlysis based on three-dimensional structure of Helicobacter pylori hp0169 gene [J]. Journal of Jilin University(Medicine Edition), 2024, 50(3): 739-748. |
| [10] | Zijia ZHU,Xia CHEN,Man CUI,Jihong WEN,Ping WANG,Dong SONG. Bioinformatics and molecular docking technology analysis on mechanism of salidroside on key differential genes of triple negative breast cancer [J]. Journal of Jilin University(Medicine Edition), 2024, 50(3): 759-769. |
| [11] | Yuting LIU,Ying YU,Guizhen LI,Qinxue SHI,Binbin LI. Bioinformatics analysis based on expression of splicing factor SRSF9 in head and neck squamous cell carcinoma and clinical significance [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 379-391. |
| [12] | Liping CHEN,Li HAN,Hua BIAN,Liye PANG. Bioinformatics analysis based on differentially expressed genes and screening of traditional Chinese medicine for treatment of severe bronchial asthma [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 411-421. |
| [13] | Minqi NING,Yong HE,Bingshu LI,Guotao HUANG,Xiaohu ZUO,Zhihan ZHAO,Wuyue HAN,Li HONG. Bioinformatics analysis based on pelvic organ prolapse related aging genes of GEO Database and LASSO regression algorithm [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 178-187. |
| [14] | Zixu YANG,Chang SU,Boyuan WANG,Chong LIU,Minghe LI. Bioinformatics analysis on molecular subtypes and clinical characteristics of head and neck squamous cell carcinoma based on genes associated with lactate metabolism [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 198-207. |
| [15] | Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252. |
|
||