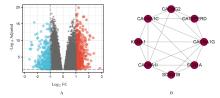
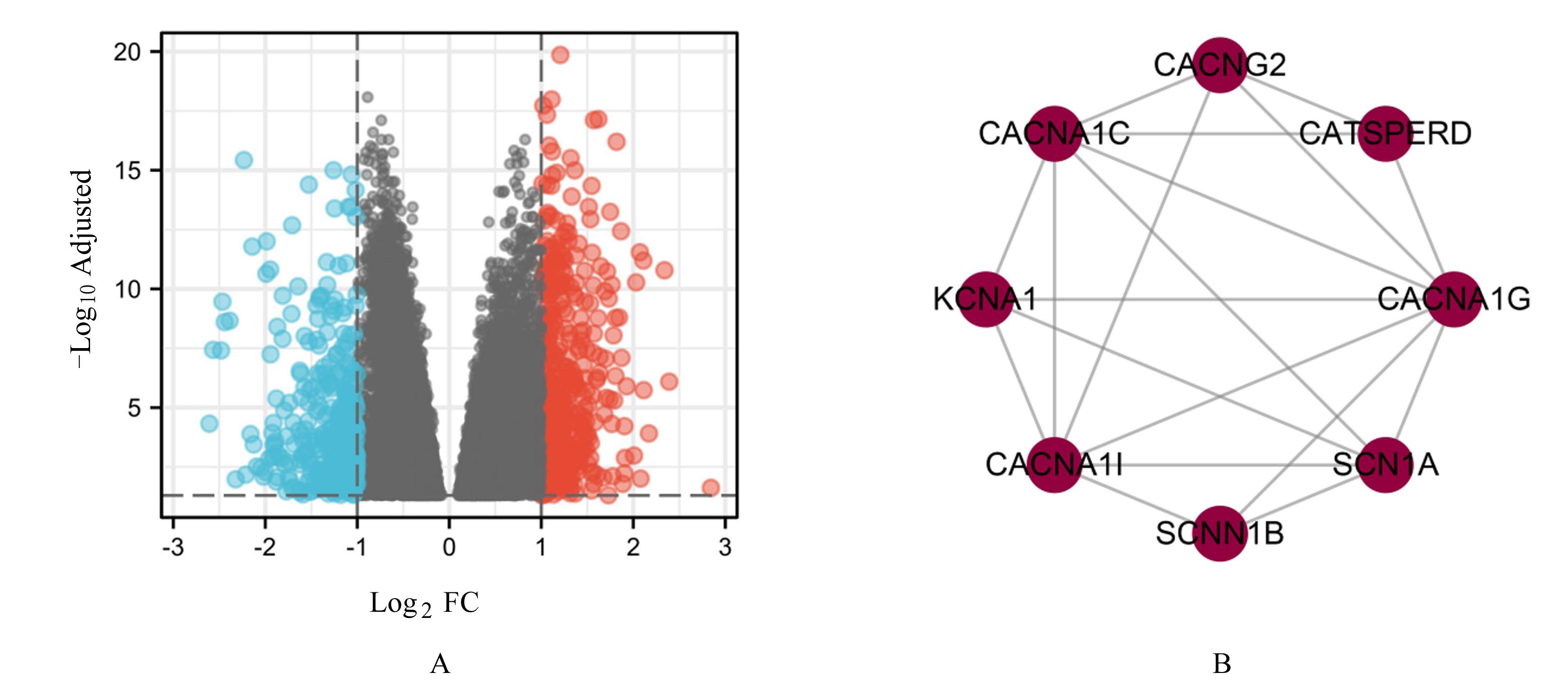
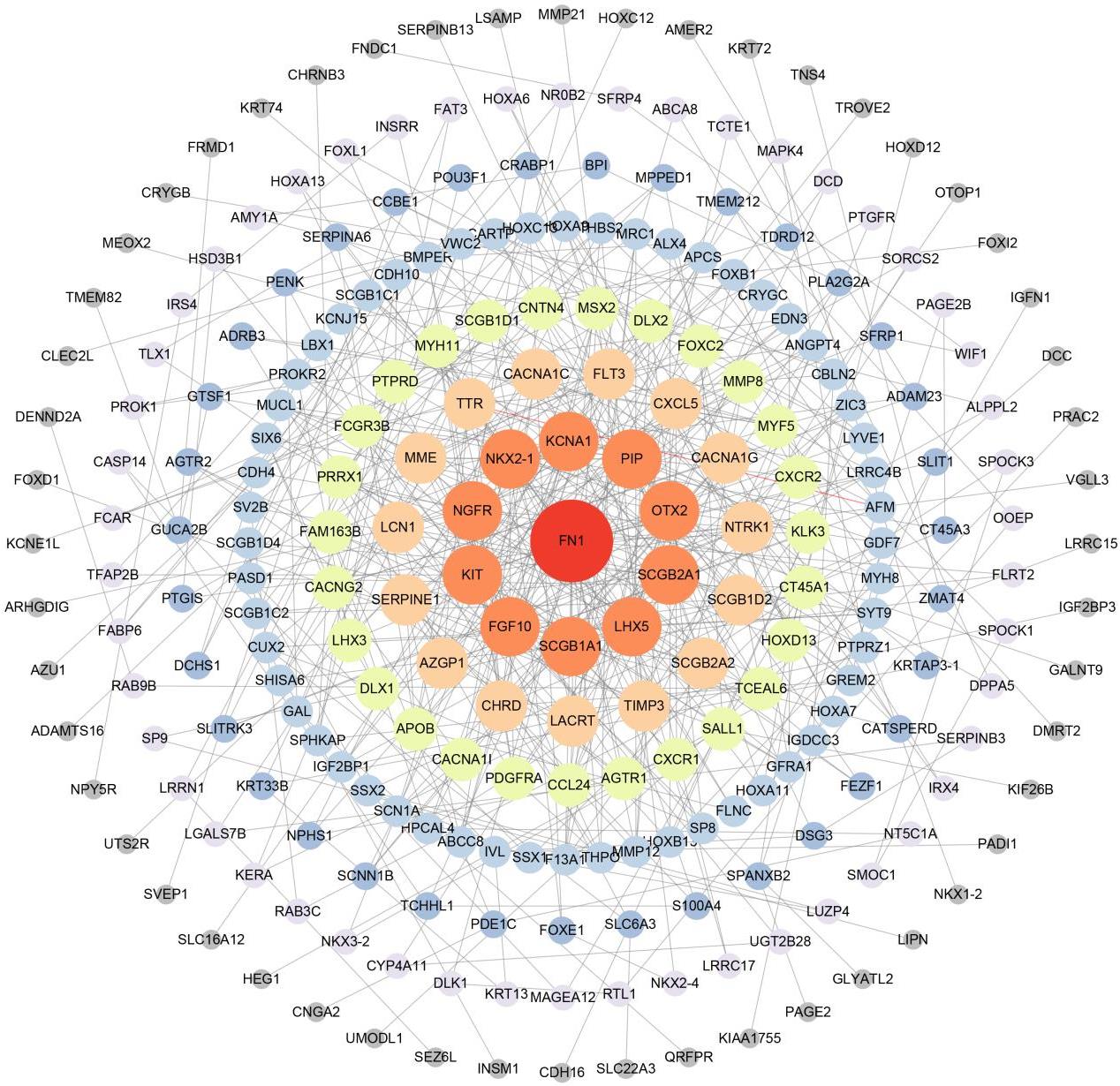
Journal of Jilin University(Medicine Edition) ›› 2023, Vol. 49 ›› Issue (2): 440-451.doi: 10.13481/j.1671-587X.20230220
• Research in clinical medicine • Previous Articles Next Articles
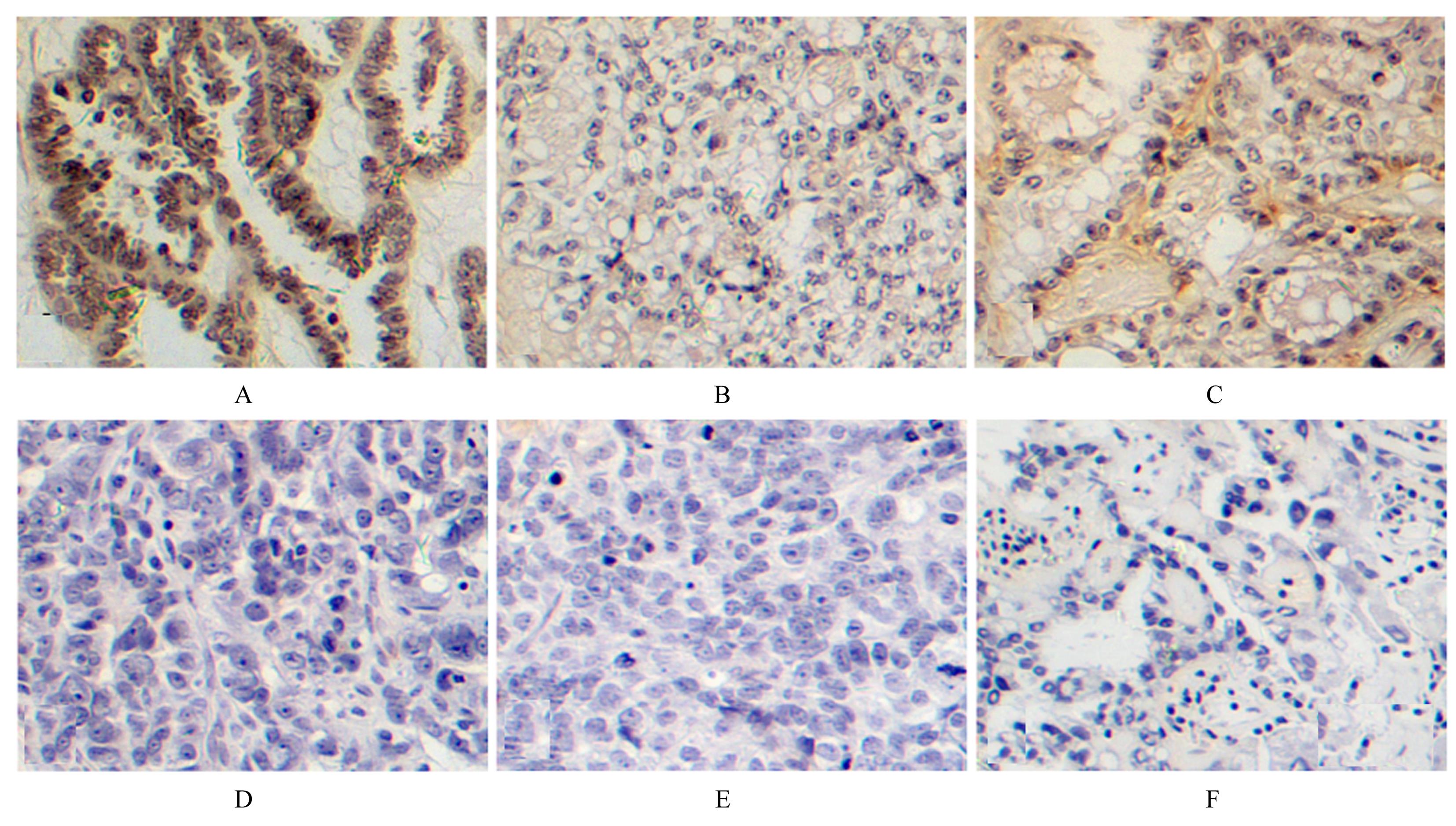
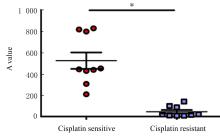
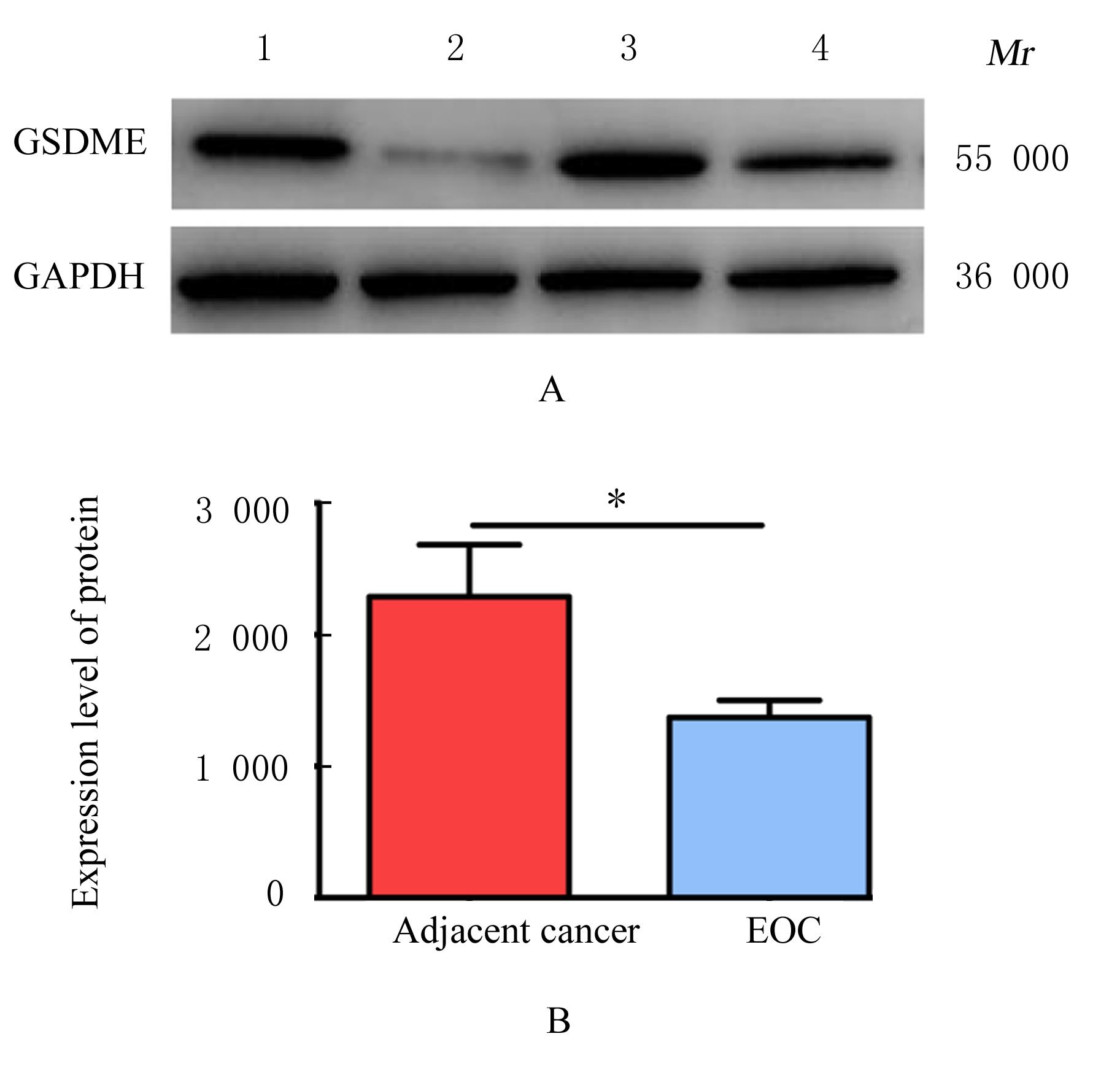
Expression level of gasdermin E in epithelial ovarian cancer tissue and its clinical significance
Mengmeng CHEN1,Junyu CHEN2,Yan GAO1,Zhengxuan FANG3,Shuhua ZHAO2,Jingying ZHENG2( ),Shuxiang LIU1(
),Shuxiang LIU1( )
)
- 1.Department of Rehabilitation Therapy,School of Nursing,Jilin University,Changchun 130021,China
2.Department of Obstetrics and Gynecology,Second Hospital,Jilin University,Changchun 130021,China
3.Department of Phamaceutics,School of Pharmacy,Yanbian University,Yanji 133002,China
-
Received:2022-08-07Online:2023-03-28Published:2023-04-24 -
Contact:Jingying ZHENG,Shuxiang LIU E-mail:zheng_jy@jlu.edu.cn;shuxiang@jlu.edu.cn
CLC Number:
- R737.31
Cite this article
Mengmeng CHEN,Junyu CHEN,Yan GAO,Zhengxuan FANG,Shuhua ZHAO,Jingying ZHENG,Shuxiang LIU. Expression level of gasdermin E in epithelial ovarian cancer tissue and its clinical significance[J].Journal of Jilin University(Medicine Edition), 2023, 49(2): 440-451.
share this article
Tab. 1
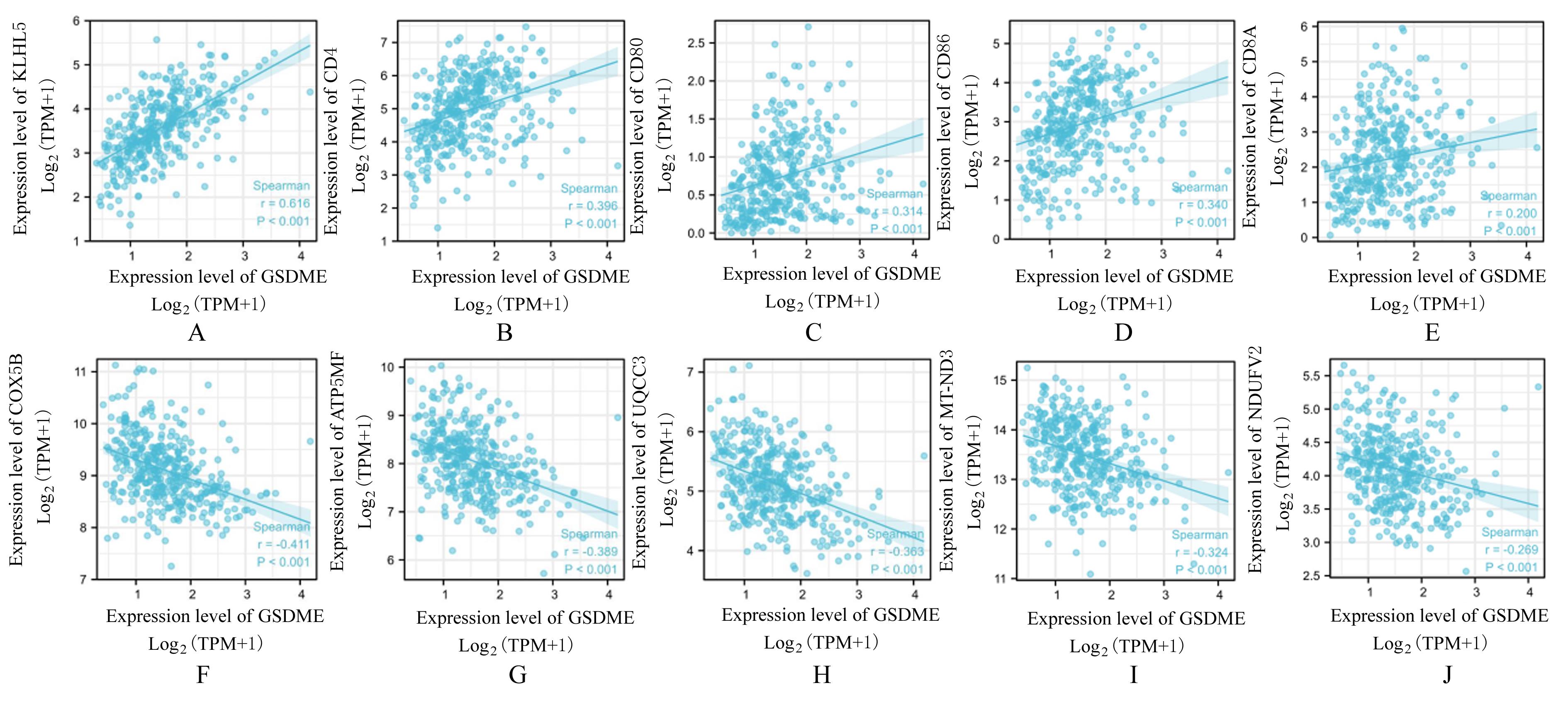
GSEA gene sets positively correlated with GSDME expression"
| ID | ES | NES | P | FDR |
|---|---|---|---|---|
| REACTOME_GPCR_LIGAND_BINDING | 0.553 | 0.652 | 0.009 | 0.005 |
| REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | 0.702 | 3.418 | 0.009 | 0.005 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 0.566 | 2.733 | 0.009 | 0.005 |
| REACTOME_ANTI_INFLAMMATORY_RESPONSE_FAVOURING_ LEISHMANIA_PARASITE_INFECTION | 0.630 | 2.963 | 0.009 | 0.005 |
| REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_ LYMPHOID_AND_A_NON_LYMPHOID_CELL | 0.652 | 3.010 | 0.009 | 0.005 |
Tab. 2
GSEA gene sets negatively correlated with GSDME expression"
| ID | ES | NES | P | FDR |
|---|---|---|---|---|
| WP_ELECTRON_TRANSPORT_CHAIN_OXPHOS_SYSTEM_IN_MITOCHONDRIA | -0.748 | -3.115 | 0.009 | 0.005 |
| WP_CYTOPLASMIC_RIBOSOMAL_PROTEINS | -0.838 | -3.454 | 0.009 | 0.005 |
| REACTOME_RRNA_PROCESSING | -0.672 | -3.145 | 0.009 | 0.005 |
| REACTOME_SIGNALING_BY_ROBO_RECEPTORS | -0.555 | -2.611 | 0.009 | 0.005 |
| REACTOME_TRANSLATION | -0.628 | -3.094 | 0.009 | 0.005 |
Tab. 3
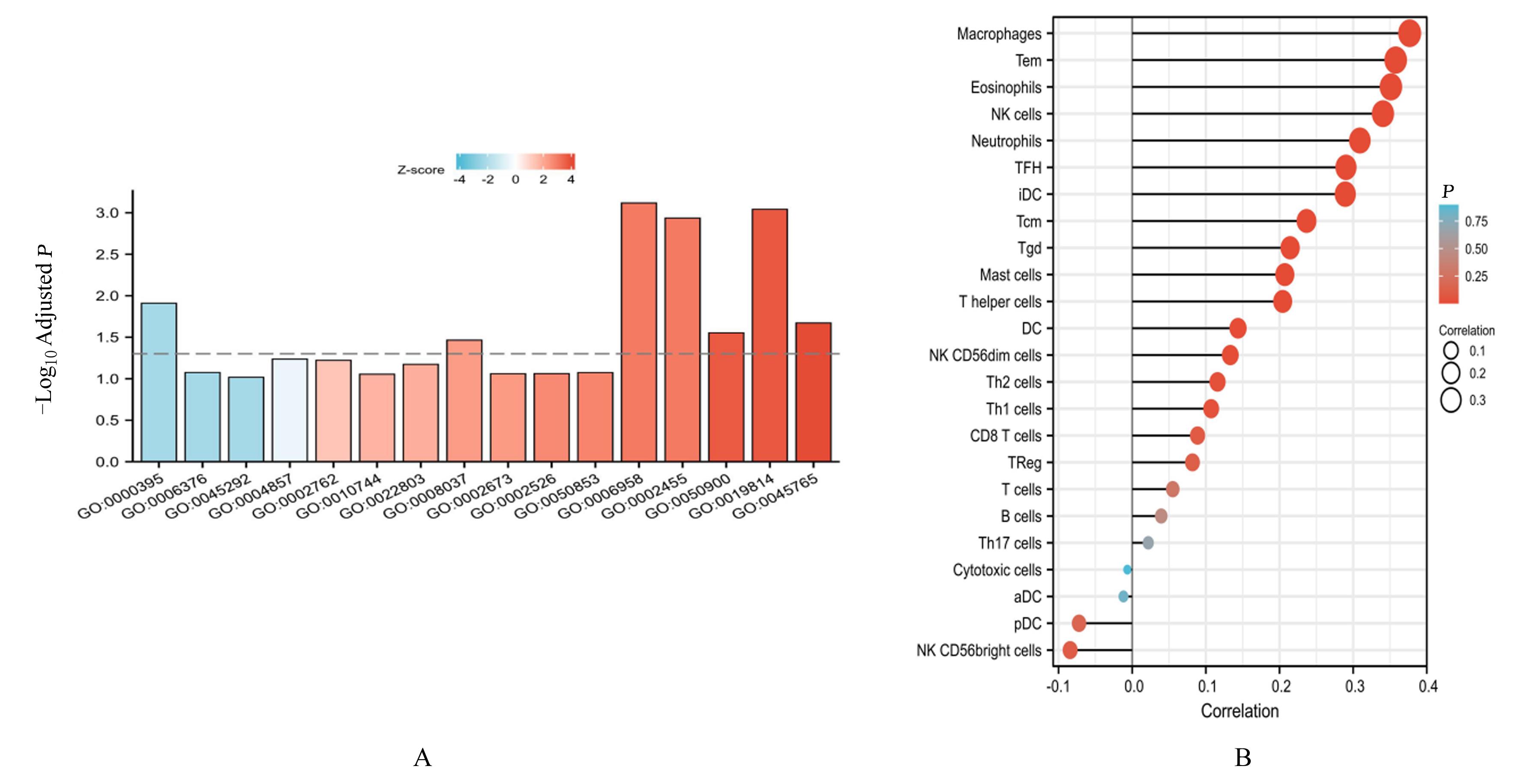
Results of GO/KEGG enrichment analysis"
| Ontology | ID | Description | Gene ratio | BgRatio | Adjusted P | q |
|---|---|---|---|---|---|---|
| BP | GO:0000395 | mRNA 5'-splice site recognition | 5/359 | 29/18 670 | 0.012 | 0.011 |
| BP | GO:0006376 | mRNA splice site selection | 5/359 | 53/18 670 | 0.084 | 0.076 |
| BP | GO:0045292 | mRNA cis splicing, via spliceosome | 5/359 | 56/18 670 | 0.096 | 0.087 |
| BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 13/359 | 150/18 670 | 0.001 | 0.001 |
| BP | GO:0050900 | leukocyte migration | 21/359 | 499/18 670 | 0.028 | 0.025 |
| CC | GO:0019814 | immunoglobulin complex | 14/387 | 159/19 717 | 0.001 | 0.001 |
| BP | GO:0045765 | regulation of angiogenesis | 18/359 | 383/18 670 | 0.021 | 0.019 |
Tab. 4
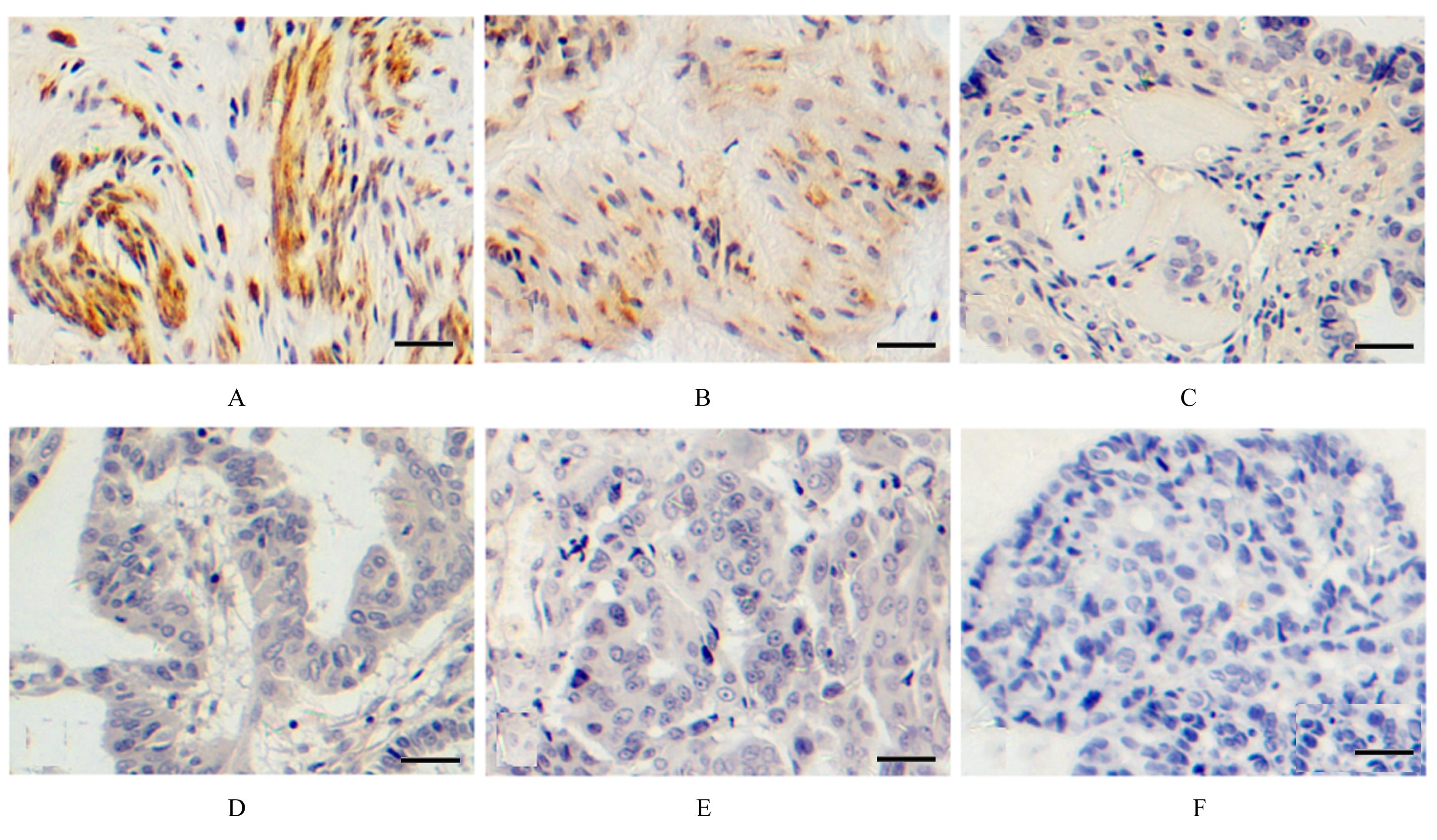
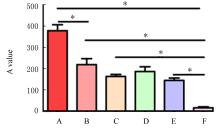
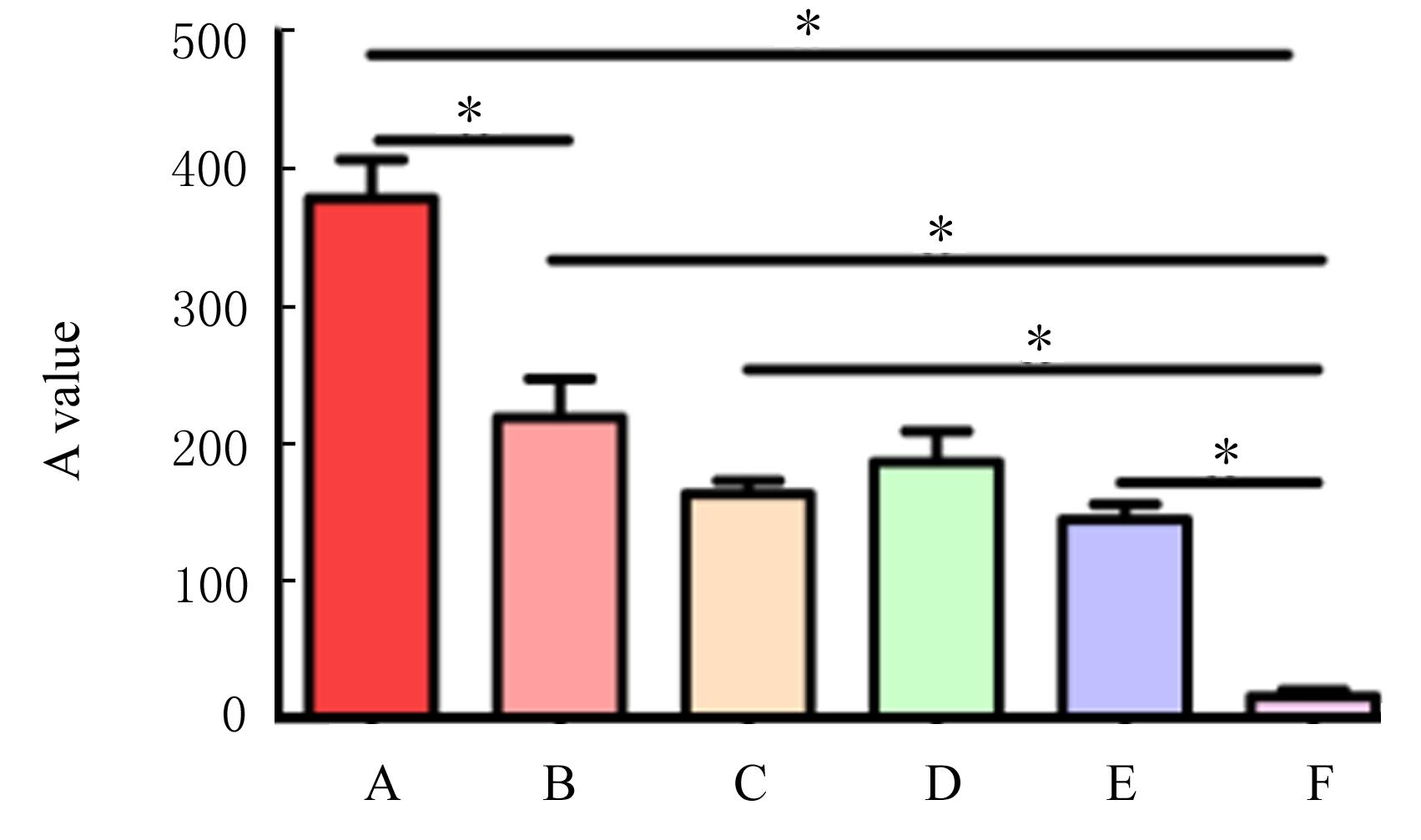
Relationship between GSDME protein expression and clinical characteristics of EOC"
| Clinical feature | n (η/%) | GSDME | χ2 | P | |
|---|---|---|---|---|---|
| Positive expression | Negative expression | ||||
| Age(year) | |||||
| <53 | 17(42.5) | 8 | 9 | ||
| ≥53 | 23(57.5) | 10 | 13 | 0.051 | 0.822 |
| FIGO stage | |||||
| Ⅰ-Ⅱ | 19(47.5) | 9 | 10 | ||
| Ⅲ-Ⅳ | 21(52.5) | 9 | 12 | 0.082 | 0.775 |
| Tumor size | |||||
| ≤10 cm | 19(47.5) | 8 | 11 | ||
| >10 cm | 21(52.5) | 10 | 11 | 0.123 | 0.726 |
| EOC type | |||||
| Type Ⅰ | 21(52.5) | 10 | 11 | ||
| Type Ⅱ | 19(47.5) | 8 | 11 | 0.123 | 0.726 |
| Pathological grade | |||||
| Low level | 16(40.0) | 11 | 5 | ||
| High level | 24(60.0) | 7 | 17 | 6.077 | 0.014 |
| Ascites volume | |||||
| <500 mL | 21(52.5) | 11 | 10 | ||
| ≥500 mL | 19(47.5) | 7 | 12 | 0.973 | 0.324 |
| 1 | LHEUREUX S, GOURLEY C, VERGOTE I, et al. Epithelial ovarian cancer[J]. Lancet,2019,393(10177): 1240-1253. |
| 2 | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J].CA Cancer J Clin,2021,71(3):209-249. |
| 3 | BARNETT R. Ovarian cancer[J]. Lancet, 2016, 387(10025): 1265. |
| 4 | MUSTAZZOLU A, BORRONI E, CIRILLO D M, et al. Trend in rifampicin-, multidrug- and extensively drug-resistant tuberculosis in Italy, 2009-2016[J]. Eur Respir J, 2018, 52(1): 1800070. |
| 5 | BERKEL C, CACAN E. Differential expression and copy number variation of gasdermin (GSDM) family members, pore-forming proteins in pyroptosis, in normal and malignant serous ovarian tissue[J]. Inflammation, 2021, 44(6): 2203-2216. |
| 6 | ZHENG Y, YUAN D, ZHANG F, et al. A systematic pan-cancer analysis of the gasdermin (GSDM) family of genes and their correlation with prognosis, the tumor microenvironment, and drug sensitivity[J]. Front Genet, 2022, 13: 926796. |
| 7 | ZHANG X W, ZHANG P, AN L, et al. Miltirone induces cell death in hepatocellular carcinoma cell through GSDME-dependent pyroptosis[J]. Acta Pharm Sin B, 2020, 10(8): 1397-1413. |
| 8 | ZHANG Z B, ZHANG Y, XIA S Y, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity[J]. Nature, 2020, 579(7799): 415-420. |
| 9 | WANG Y B, YIN B, LI D N, et al. GSDME mediates caspase-3-dependent pyroptosis in gastric cancer[J]. Biochem Biophys Res Commun,2018,495(1):1418-1425. |
| 10 | TAN G, HUANG C Y, CHEN J Y, et al. HMGB1 released from GSDME-mediated pyroptotic epithelial cells participates in the tumorigenesis of colitis-associated colorectal cancer through the ERK1/2 pathway[J]. J Hematol Oncol, 2020, 13(1): 149. |
| 11 | 石 瑛, 任静静, 梁 晨, 等. GSDME通过调控细胞焦亡影响乳腺癌MCF-7细胞对紫杉醇的敏感性[J]. 中国肿瘤生物治疗杂志, 2019, 26(2): 146-151. |
| 12 | 高世华, 李植锋, 蔡键锋. GSDME通过p53、caspase3增强小细胞肺癌细胞的紫杉醇敏感[J]. 医学研究杂志, 2021, 50(10): 138-142. |
| 13 | JIANG M X, QI L, LI L S,et al.The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer[J]. Cell Death Discov, 2020, 6: 112. |
| 14 | GALLET P, OUSSALAH A, POUGET C, et al. Integrative genomics analysis of nasal intestinal-type adenocarcinomas demonstrates the major role of CACNA1C and paves the way for a simple diagnostic tool in male woodworkers[J]. Clin Epigenetics, 2021, 13(1): 179. |
| 15 | KRAUS D, WEIDER S, PROBSTMEIER R, et al. Neoexpression of JUNO in oral tumors is accompanied with the complete suppression of four other genes and suggests the application of new biomarker tools[J]. J Pers Med, 2022, 12(3): 494. |
| 16 | TONMOY M I Q, FARIHA A, HAMI I, et al. Computational epigenetic landscape analysis reveals association of CACNA1G-AS1,F11-AS1,NNT-AS1,and MSC-AS1 lncRNAs in prostate cancer progression through aberrant methylation[J]. Sci Rep, 2022,12(1): 10260. |
| 17 | YAN Y, HE W, CHEN Y, et al. Comprehensive analysis to identify the encoded gens of sodium channels as a prognostic biomarker in hepatocellular carcinoma[J]. Front Genet, 2022, 12: 802067. |
| 18 | MARIANI A, WANG C, OBERG A L, et al. Genes associated with bowel metastases in ovarian cancer[J]. Gynecol Oncol, 2019, 154(3): 495-504. |
| 19 | PENG C J, LI L C, LUO G X, et al. Integrated analysis of the M2 macrophage-related signature associated with prognosis in ovarian cancer[J]. Front Oncol, 2022, 12: 986885. |
| 20 | ZHAO W P, WANG H W, LIU J, et al. Mitochondrial respiratory chain complex abnormal expressions and fusion disorder are involved in fluoride-induced mitochondrial dysfunction in ovarian granulosa cells[J]. Chemosphere, 2019, 215: 619-625. |
| 21 | ANNESLEY S J, FISHER P R. Mitochondria in health and disease[J]. Cells, 2019, 8(7): 680. |
| 22 | ASHRAFI G, SCHWARZ T L. The pathways of mitophagy for quality control and clearance of mitochondria[J]. Cell Death Differ, 2013,20(1): 31-42. |
| 23 | SHARMA P, HU-LIESKOVAN S, WARGO J A, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy[J]. Cell, 2017,168(4): 707-723. |
| 24 | LI F, XIA Q, REN L, et al. GSDME increases chemotherapeutic drug sensitivity by inducing pyroptosis in retinoblastoma cells[J]. Oxid Med Cell Longev, 2022, 2022: 2371807. |
| 25 | RIOJA-BLANCO E, ARROYO-SOLERA I, ÁLAMO P, et al. CXCR4-targeted nanotoxins induce GSDME-dependent pyroptosis in head and neck squamous cell carcinoma[J]. J Exp Clin Cancer Res, 2022, 41(1): 49. |
| [1] | Xiaoyan WANG,Yihong HU,Yucheng HAN,Xianqiong ZOU. Bioinformatics analysis on mechanism of COMMD7 in occurrence and development of brain low-grade glioma [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 414-424. |
| [2] | Rui WANG,Ding ZHANG,Ruijian ZHUGE,Qian XUE,Li GUO. Bioinformatics analysis on mechanism of liver injury induced by hexavalent chromium [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 452-459. |
| [3] | Zhiyuan XIAO,Bing SONG,Xinyu MA,Lianhui JIN,Tong ZHENG,Fang CHAI. Bioinformatics analysis on expression of apoptosis related gene CD44 in thyroid carcinoma tissue and its relationship with tumor invasion and immune cell infiltration [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 473-481. |
| [4] | Wuyue TANG,Shuojie LI,Lijuan PANG. Bioinformatics analysis of splicing factor-alternative splicing regulatory network based on alternative splicing data in lung adenocarcinoma tissue [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 139-149. |
| [5] | Jilin FAN,Tingting ZHU,Xiaoling TIAN,Sijia LIU,Jing SU,Shiliang ZHANG. Bioinformatics analysis based on circRNA-miRNA-mRNA network construction and immune cell infiltration in atrial fibrillation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1535-1545. |
| [6] | Xiaoyan WANG,Qiuyue ZHANG,Yujie HU,Zhijing MO. Bioinformatics analysis based on molecular characteristics and mechanism of cystic fibrosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1290-1297. |
| [7] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
| [8] | Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765. |
| [9] | Xiaoyan LI,Wei ZHANG,Jie HE. Promotion effect of REG1A on proliferation and migration of lung adenocarcinoma cells by regulating Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 444-453. |
| [10] | Ying ZHAO,Danyu ZHAO,Chao LIU. Bioinformatics analysis on prognostic evaluation value of TXNDC11 gene in pan-cancer and its immunity regulation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 142-153. |
| [11] | Zhijuan ZHAO,Lian MENG,Chunxia LIU. Bioinformatics analysis on miRNA-mRNA regulatory networks based on fusion genes acting in rhabdomyosarcoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 154-162. |
| [12] | Nan LI,Lei CHEN,Tianmin XU,Kun ZHANG. Bioinformatics analysis on screening of extracellular matrix related genes in patients with endometriosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 188-194. |
| [13] | Hao DONG,Haizhen JIANG,Chao LI,Dengke GAO,Yaping JIN,Huatao CHEN. Construction of mouse NR1D1 gene overexpression vector and its bioinformatics analysis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(1): 94-103. |
| [14] | Jue CHEN,Li HONG,Suting LI,Zhi WANG,Mao CHEN,Menglei HAO,Ya XIAO,Jie MIN,Ming HU. Expressions of vesicle transport-related genes during repair of pelvic floor muscles after injury [J]. Journal of Jilin University(Medicine Edition), 2021, 47(5): 1092-1098. |
| [15] | Yang YU,Sainan LIU,Yunkai LIU,Yong LI,Yichun QIAO,Yi CHENG. Bioinformatic analysis on expression characteristics of PRPS2 and its relationship with prognosis of breast cancer [J]. Journal of Jilin University(Medicine Edition), 2021, 47(5): 1229-1236. |
|
||