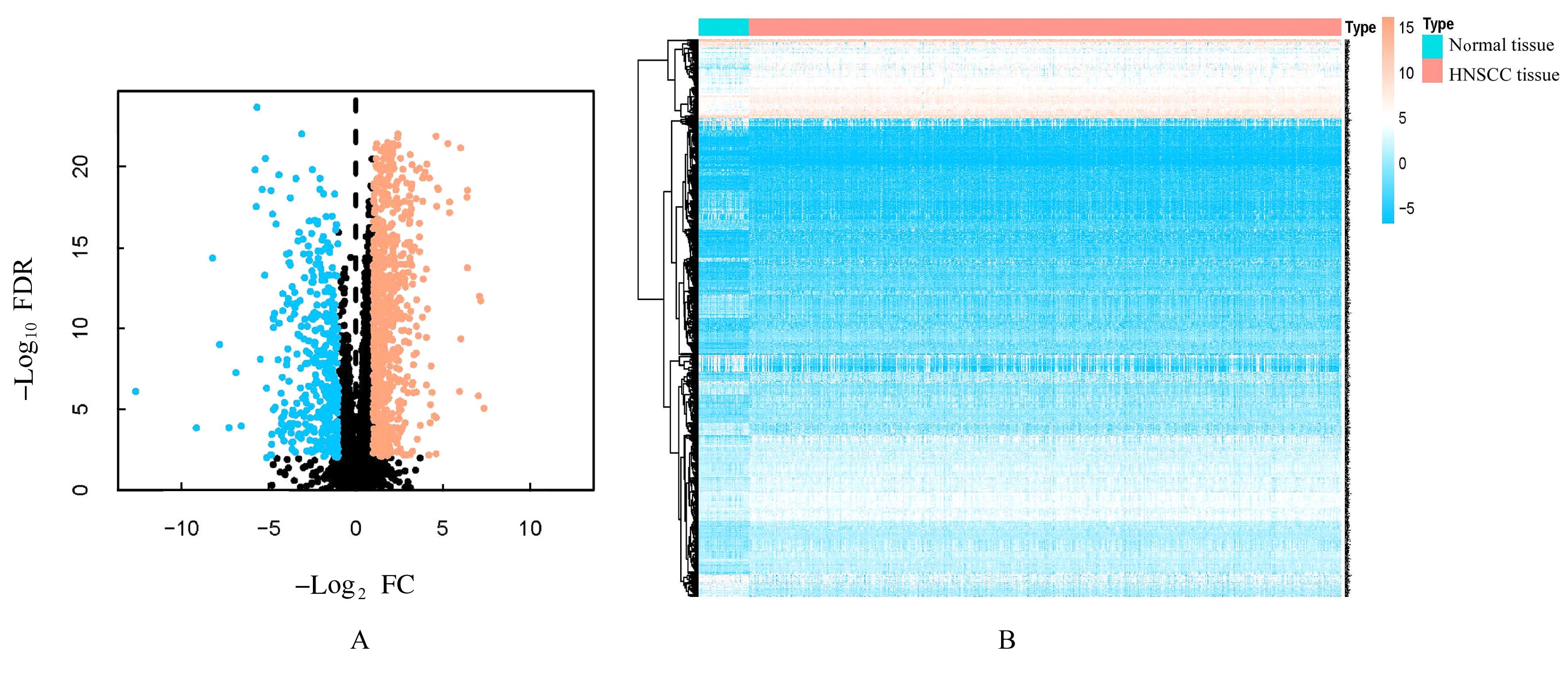
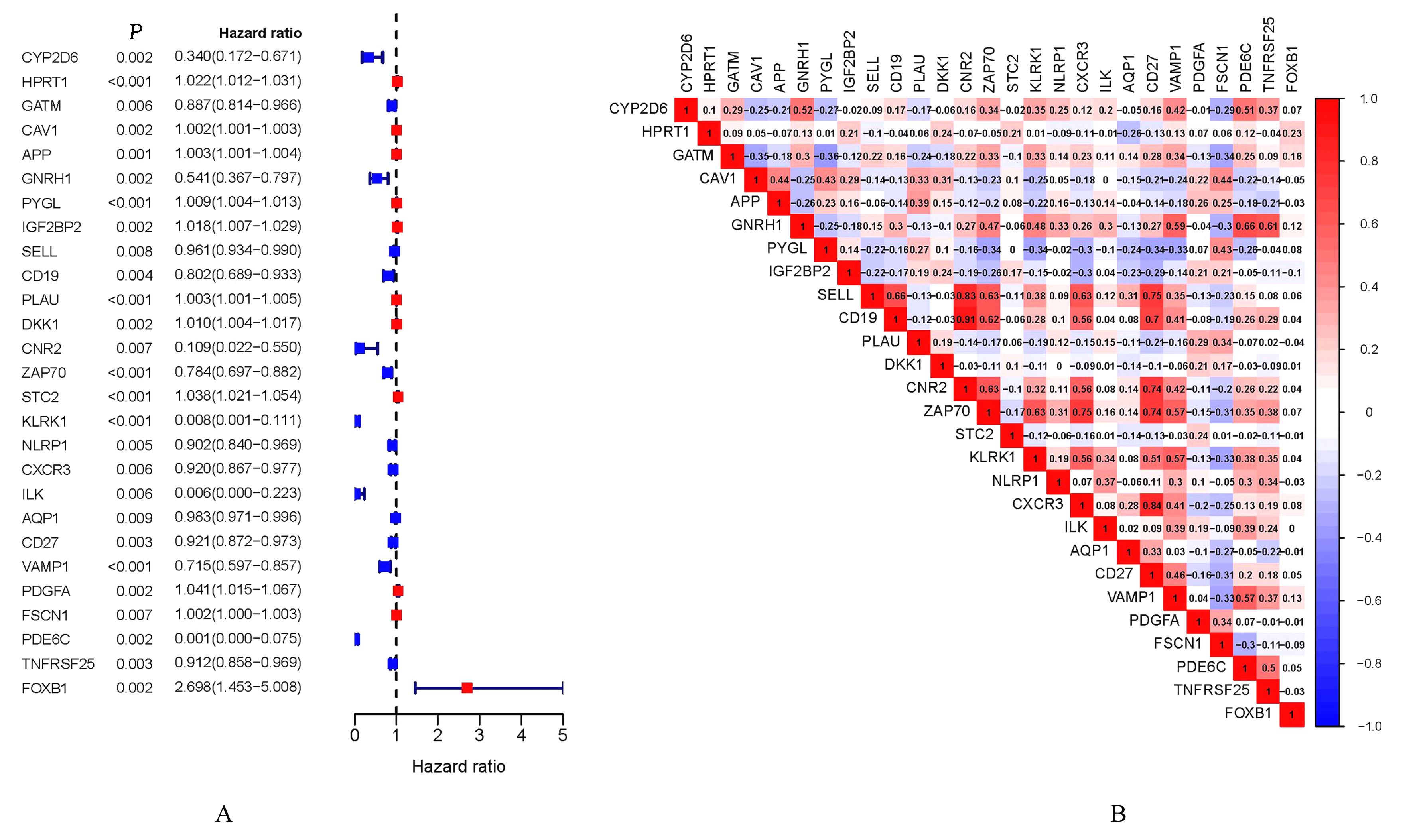
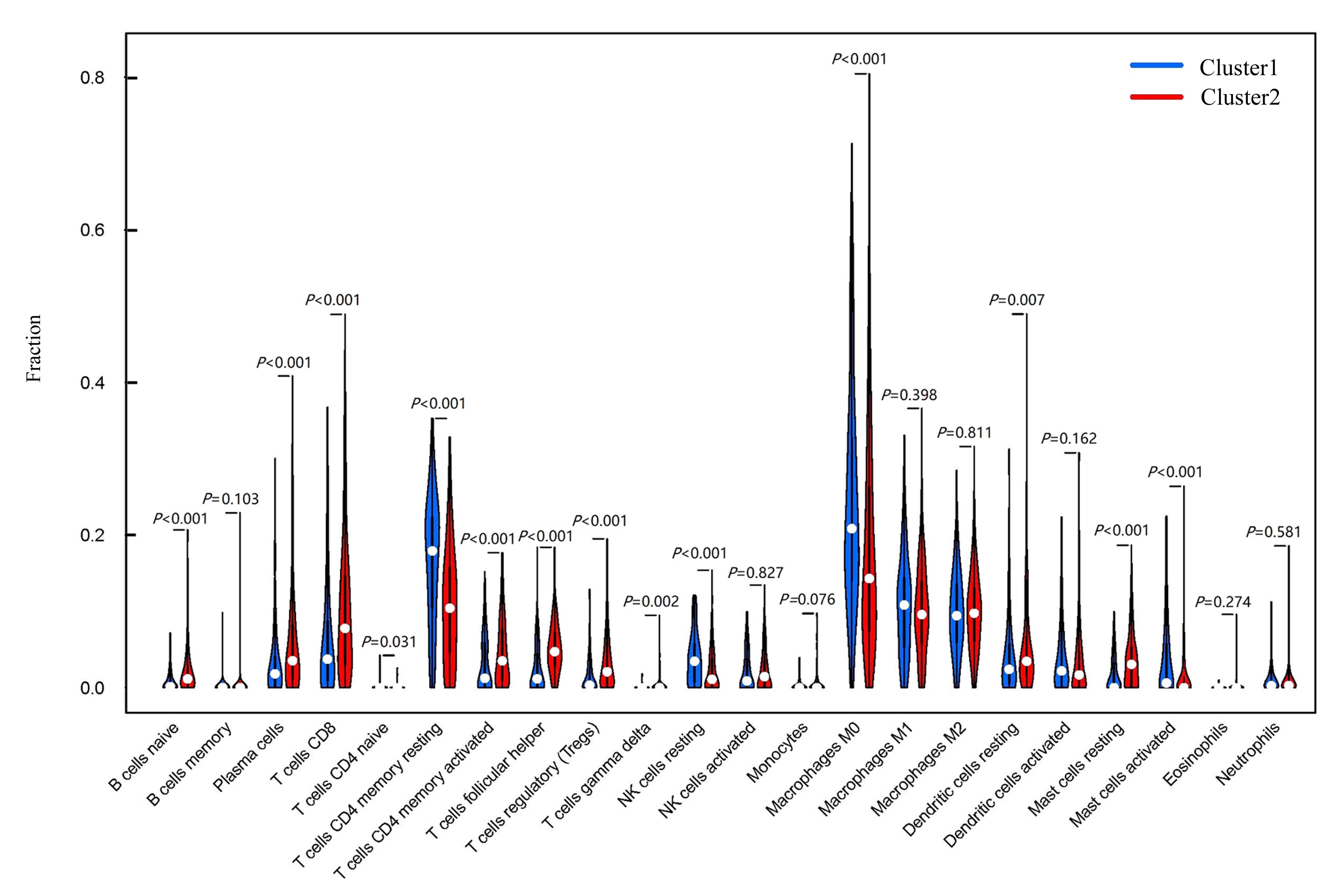
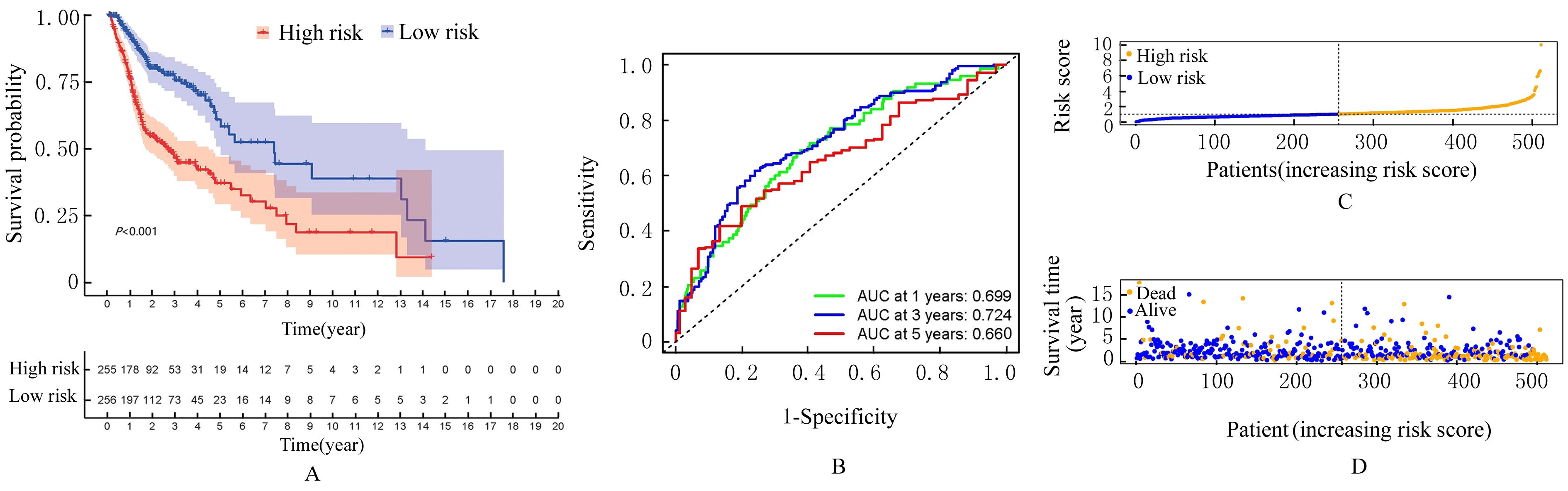
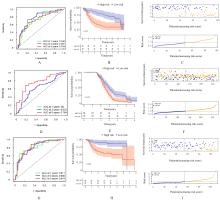
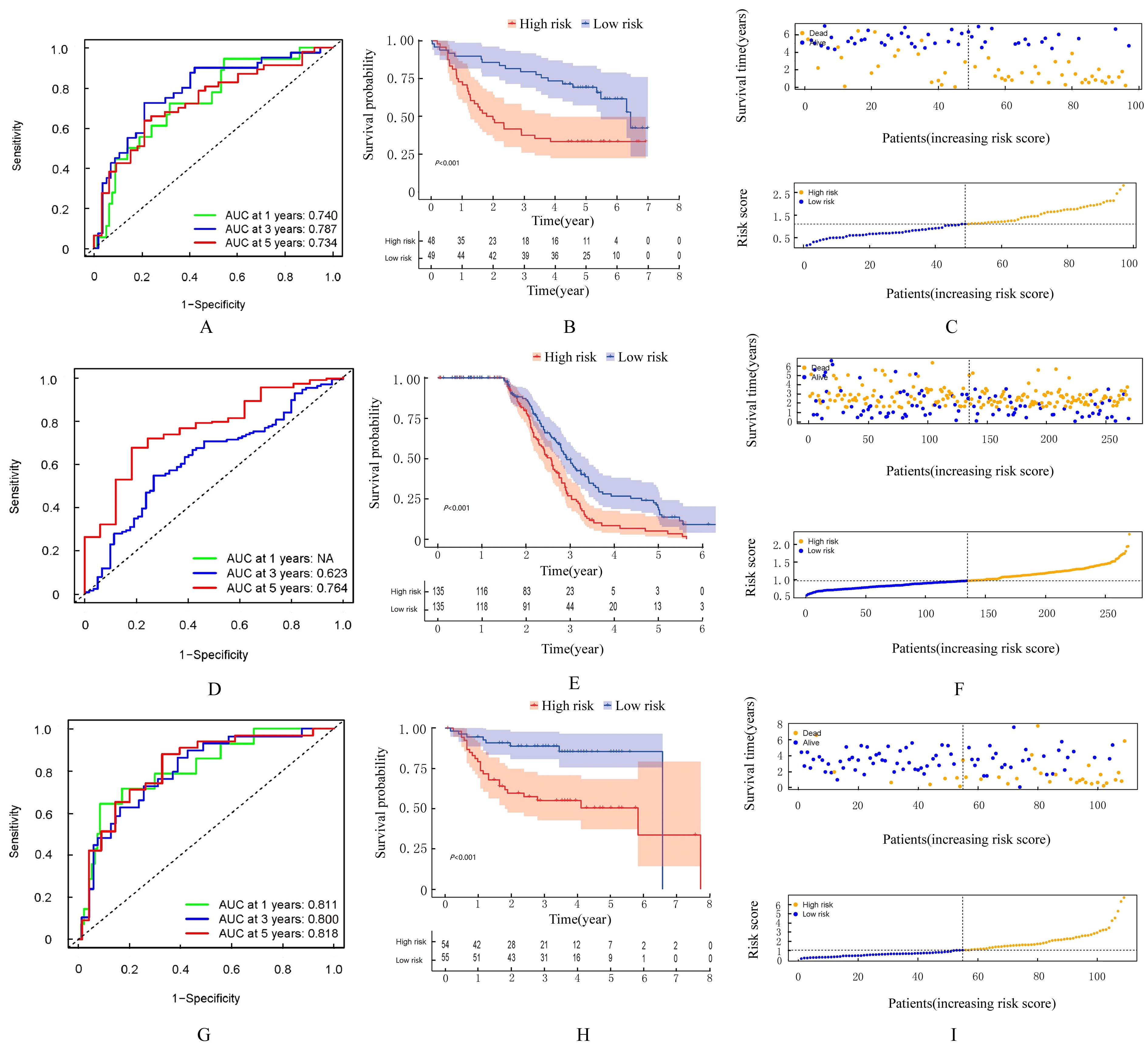
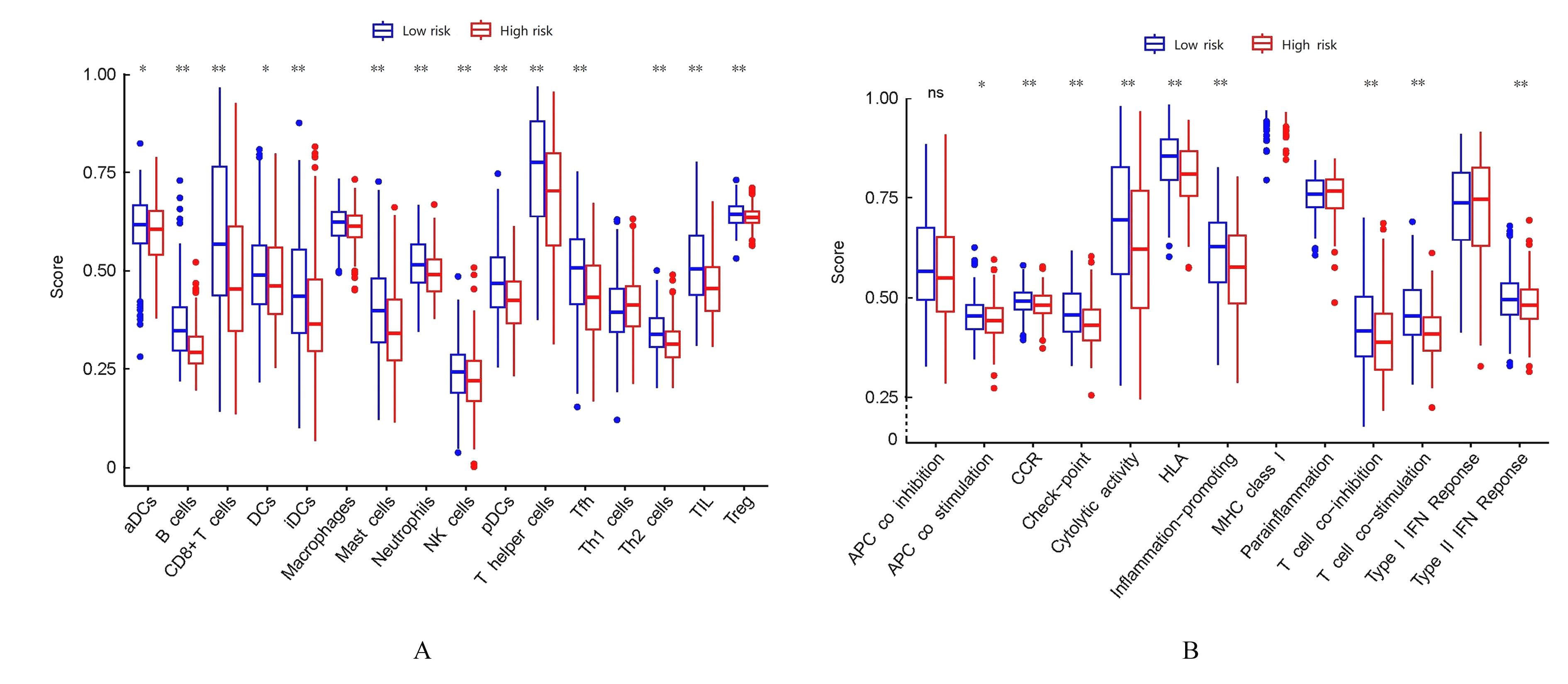
Journal of Jilin University(Medicine Edition) ›› 2024, Vol. 50 ›› Issue (1): 198-207.doi: 10.13481/j.1671-587X.20240124
• Research in clinical medicine • Previous Articles
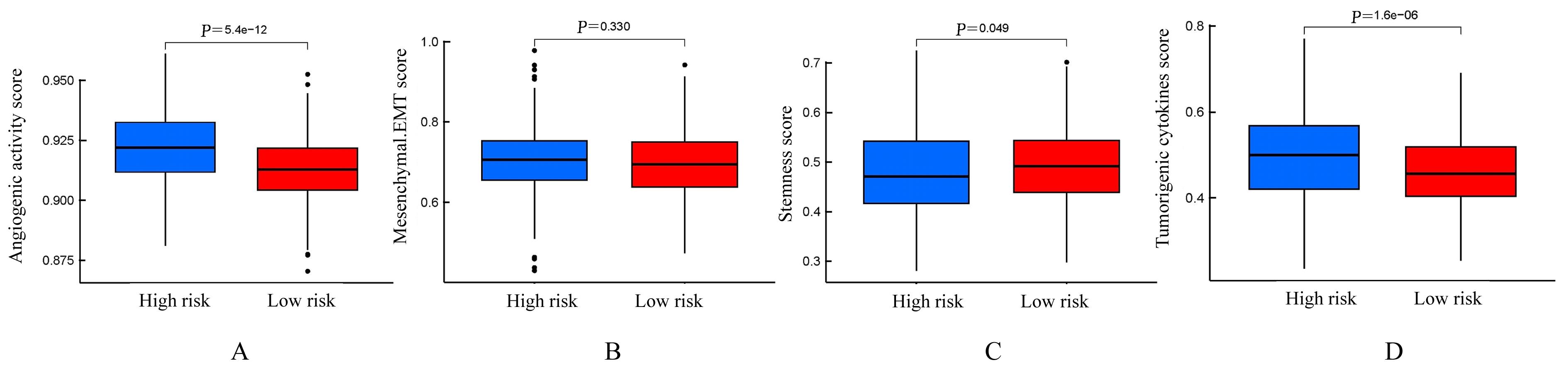
Bioinformatics analysis on molecular subtypes and clinical characteristics of head and neck squamous cell carcinoma based on genes associated with lactate metabolism
Zixu YANG1,Chang SU1,Boyuan WANG1,Chong LIU1,Minghe LI1,2( )
)
- 1.Department of Oral and Maxillofacial Surgery,Stomatology Hospital,Jilin University,Changchun 130021,China
2.Department of Stomatology,Second Hospital,Jilin University,Changchun 130022,China
-
Received:2023-04-13Online:2024-01-28Published:2024-01-31 -
Contact:Minghe LI E-mail:liminghe@jlu.edu.cn
CLC Number:
- R782
Cite this article
Zixu YANG,Chang SU,Boyuan WANG,Chong LIU,Minghe LI. Bioinformatics analysis on molecular subtypes and clinical characteristics of head and neck squamous cell carcinoma based on genes associated with lactate metabolism[J].Journal of Jilin University(Medicine Edition), 2024, 50(1): 198-207.
share this article
| 1 | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J].CA Cancer J Clin,2021,71(3):209-249. |
| 2 | NAMAYANDEH S M, KHAZAEI Z, LARI NAJAFI M, et al. GLOBAL leukemia in children 0-14 statistics 2018, incidence and mortality and human development index (HDI): GLOBOCAN sources and methods[J]. Asian Pac J Cancer Prev, 2020, 21(5): 1487-1494. |
| 3 | JOHNSON D E, BURTNESS B, LEEMANS C R,et al.Head and neck squamous cell carcinoma[J]. Nat Rev Dis Primers, 2020, 6(1): 92. |
| 4 | LEEMANS C R, BRAAKHUIS B J M, BRAKENHOFF R H. Response to correspondence on the molecular biology of head and neck cancer[J]. Nat Rev Cancer, 2011, 11(5): 382. |
| 5 | CHAUHAN S S, KAUR J, KUMAR M, et al. Prediction of recurrence-free survival using a protein expression-based risk classifier for head and neck cancer[J]. Oncogenesis, 2015, 4(4): e147. |
| 6 | CHEN Y J, MAHIEU N G, HUANG X J, et al. Lactate metabolism is associated with mammalian mitochondria[J].Nat Chem Biol,2016,12(11): 937-943. |
| 7 | FAUBERT B, LI K Y, CAI L, et al. Lactate metabolism in human lung tumors[J]. Cell, 2017,171(2): 358-371.e9. |
| 8 | CHEN L H, HUANG L X, GU Y,et al.Lactate-lactylation hands between metabolic reprogramming and immunosuppression[J]. Int J Mol Sci, 2022,23(19): 11943. |
| 9 | MAI S J, LIANG L P, MAI G H, et al. Development and validation of lactate metabolism-related lncRNA signature as a prognostic model for lung adenocarcinoma[J]. Front Endocrinol,2022,13: 829175. |
| 10 | SUN Z L, TAO W, GUO X D, et al. Construction of a lactate-related prognostic signature for predicting prognosis, tumor microenvironment, and immune response in kidney renal clear cell carcinoma[J]. Front Immunol, 2022, 13: 818984. |
| 11 | ZHANG Z H, FANG T, LV Y G. A novel lactate metabolism-related signature predicts prognosis and tumor immune microenvironment of breast cancer[J]. Front Genet, 2022, 13: 934830. |
| 12 | FISHILEVICH S, NUDEL R, RAPPAPORT N,et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards[J]. Database, 2017, 2017: bax028. |
| 13 | HIRSCHHAEUSER F, SATTLER U G, MUELLER-KLIESER W. Lactate: a metabolic key player in cancer[J].Cancer Res,2011,71(22):6921-6925. |
| 14 | CHALMERS Z R, CONNELLY C F, FABRIZIO D, et al. Analysis of 100, 000 human cancer genomes reveals the landscape of tumor mutational burden[J]. Genome Med, 2017, 9(1): 34. |
| 15 | MARABELLE A, FAKIH M, LOPEZ J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study[J]. Lancet Oncol, 2020, 21(10): 1353-1365. |
| 16 | WU T, JIAO Z, LI Y X, et al. HPRT1 promotes chemoresistance in oral squamous cell carcinoma via activating MMP1/PI3K/akt signaling pathway[J]. Cancers, 2022, 14(4): 855. |
| 17 | JIANG Z H, SHEN X F, WEI Y H,et al. A pan-cancer analysis reveals the prognostic and immunotherapeutic value of stanniocalcin-2 (STC2)[J]. Front Genet, 2022, 13: 927046. |
| 18 | XIE W, LI X Y, YANG C X, et al. The pyroptosis-related gene prognostic index associated with tumor immune infiltration for pancreatic cancer[J]. Int J Mol Sci, 2022, 23(11): 6178. |
| 19 | BURR M L, SPARBIER C E, CHAN K L, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer[J]. Cancer Cell, 2019, 36(4): 385-401.e8. |
| 20 | GUO X Y, LIN W Q, WEN W Q, et al. Identifying novel susceptibility genes for colorectal cancer risk from a transcriptome-wide association study of 125,478 subjects[J]. Gastroenterology, 2021, 160(4): 1164-1178. |
| 21 | XU J W, LIU X Y, LIU X T, et al. Long noncoding RNA KCNMB2-AS1 promotes the development of esophageal cancer by modulating the miR-3194-3p/PYGL axis[J]. Bioengineered, 2021, 12(1):6687-6702. |
| 22 | GAO Y, ZHANG E C, FEI X, et al. Identification of novel metabolism-associated subtypes for pancreatic cancer to establish an eighteen-gene risk prediction model[J]. Front Cell Dev Biol, 2021, 9: 691161. |
| 23 | ZHONG H, LIU S, CAO F, et al. Dissecting tumor antigens and immune subtypes of glioma to develop mRNA vaccine[J]. Front Immunol, 2021, 12: 709986. |
| 24 | WEI M Q, LI H, LI Q F, et al. Based on network pharmacology to explore the molecular targets and mechanisms of Gegen Qinlian Decoction for the treatment of ulcerative colitis[J]. Biomed Res Int, 2020, 2020: 5217405. |
| 25 | HU B, YANG X B, SANG X T. Development and verification of the hypoxia-related and immune-associated prognosis signature for hepatocellular carcinoma[J]. J Hepatocell Carcinoma, 2020, 7: 315-330. |
| 26 | WU M, WEI B, DUAN S L, et al. Methylation-driven gene PLAU as a potential prognostic marker for differential thyroid carcinoma[J]. Front Cell Dev Biol, 2022, 10: 819484. |
| 27 | AI C, ZHANG J X, LIAN S Y, et al. FOXM1 functions collaboratively with PLAU to promote gastric cancer progression[J]. J Cancer, 2020, 11(4): 788-794. |
| 28 | VISHNUBALAJI R, ALAJEZ N M. Epigenetic regulation of triple negative breast cancer (TNBC) by TGF-β signaling[J]. Sci Rep, 2021, 11(1): 15410. |
| 29 | XIONG X X, CHEN S Y, SHEN J F, et al. Cannabis suppresses antitumor immunity by inhibiting JAK/STAT signaling in T cells through CNR2[J]. Signal Transduct Target Ther, 2022, 7(1): 99. |
| 30 | EKE I, HEHLGANS S, CORDES N. There’s something about ilk[J]. Int J Radiat Biol, 2009,85(11): 929-936. |
| 31 | KATOH M, KATOH M. Human FOX gene family (Review)[J]. Int J Oncol, 2004, 25(5): 1495-1500. |
| [1] | Minqi NING,Yong HE,Bingshu LI,Guotao HUANG,Xiaohu ZUO,Zhihan ZHAO,Wuyue HAN,Li HONG. Bioinformatics analysis based on pelvic organ prolapse related aging genes of GEO Database and LASSO regression algorithm [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 178-187. |
| [2] | Hui YE,Zhe SUN,Liting ZHOU,Wen QI,Lin YE. Bioinformatics analysis on differentially expressed genes in lung adenocarcinoma based on GEO and TCGA Databases [J]. Journal of Jilin University(Medicine Edition), 2023, 49(6): 1491-1503. |
| [3] | Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252. |
| [4] | Haikang CUI,Xudong ZHANG,Xiaoning LI,Xi YANG,Lan YANG,Wenjie ZHANG. Bioinformatics analysis on predition effect of subtypes of cell pyroptosis and APOD on prognosis of gastric cancer patients [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1268-1279. |
| [5] | Yiming ZHAO,Haiyang XU. Bioinformatics analysis on related genes and candidate pathways of glioblastoma multiforme [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1280-1289. |
| [6] | Xiaoying ZHAO,Jiansheng SU,Jiahui MA,Yuefeng WANG,Dan WANG,Liyuan SUN. Bioinformatics analysis based on activity of heterotrimeric peptide H5LL and construction of recombinant Lactic acid bacteria expression vector [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1366-1374. |
| [7] | Xiaoyan WANG,Yihong HU,Yucheng HAN,Xianqiong ZOU. Bioinformatics analysis on mechanism of COMMD7 in occurrence and development of brain low-grade glioma [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 414-424. |
| [8] | Rui WANG,Ding ZHANG,Ruijian ZHUGE,Qian XUE,Li GUO. Bioinformatics analysis on mechanism of liver injury induced by hexavalent chromium [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 452-459. |
| [9] | Zhiyuan XIAO,Bing SONG,Xinyu MA,Lianhui JIN,Tong ZHENG,Fang CHAI. Bioinformatics analysis on expression of apoptosis related gene CD44 in thyroid carcinoma tissue and its relationship with tumor invasion and immune cell infiltration [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 473-481. |
| [10] | Wuyue TANG,Shuojie LI,Lijuan PANG. Bioinformatics analysis of splicing factor-alternative splicing regulatory network based on alternative splicing data in lung adenocarcinoma tissue [J]. Journal of Jilin University(Medicine Edition), 2023, 49(1): 139-149. |
| [11] | Jilin FAN,Tingting ZHU,Xiaoling TIAN,Sijia LIU,Jing SU,Shiliang ZHANG. Bioinformatics analysis based on circRNA-miRNA-mRNA network construction and immune cell infiltration in atrial fibrillation [J]. Journal of Jilin University(Medicine Edition), 2022, 48(6): 1535-1545. |
| [12] | Xiaoyan WANG,Qiuyue ZHANG,Yujie HU,Zhijing MO. Bioinformatics analysis based on molecular characteristics and mechanism of cystic fibrosis [J]. Journal of Jilin University(Medicine Edition), 2022, 48(5): 1290-1297. |
| [13] | Liantao HU,Wenjun DENG,Shizhen LU,Luo SUN,Xuebing LI,Chuhao LI,Xinran WANG,chunbing ZHANG,Yue LI,Weiqun WANG. Bioinformatics analysis on CC chemokine ligand 20 expression in hepatocellular carcinoma tissue and its effect on prognostic assessment of liver hepatocellular carcinoma [J]. Journal of Jilin University(Medicine Edition), 2022, 48(4): 1010-1017. |
| [14] | Qian LIU,Guoping QI,Huayi YU,Yuyang DAI,Wenbin LU,Jianhua JIN. Bioinformatics analysis on screening of colon cancer core genes and independent prognostic factors [J]. Journal of Jilin University(Medicine Edition), 2022, 48(3): 755-765. |
| [15] | Xiaoyan LI,Wei ZHANG,Jie HE. Promotion effect of REG1A on proliferation and migration of lung adenocarcinoma cells by regulating Wnt/β-catenin signaling pathway [J]. Journal of Jilin University(Medicine Edition), 2022, 48(2): 444-453. |
|
||