Journal of Jilin University(Medicine Edition) ›› 2024, Vol. 50 ›› Issue (4): 1076-1086.doi: 10.13481/j.1671-587X.20240422
• Research in clinical medicine • Previous Articles Next Articles
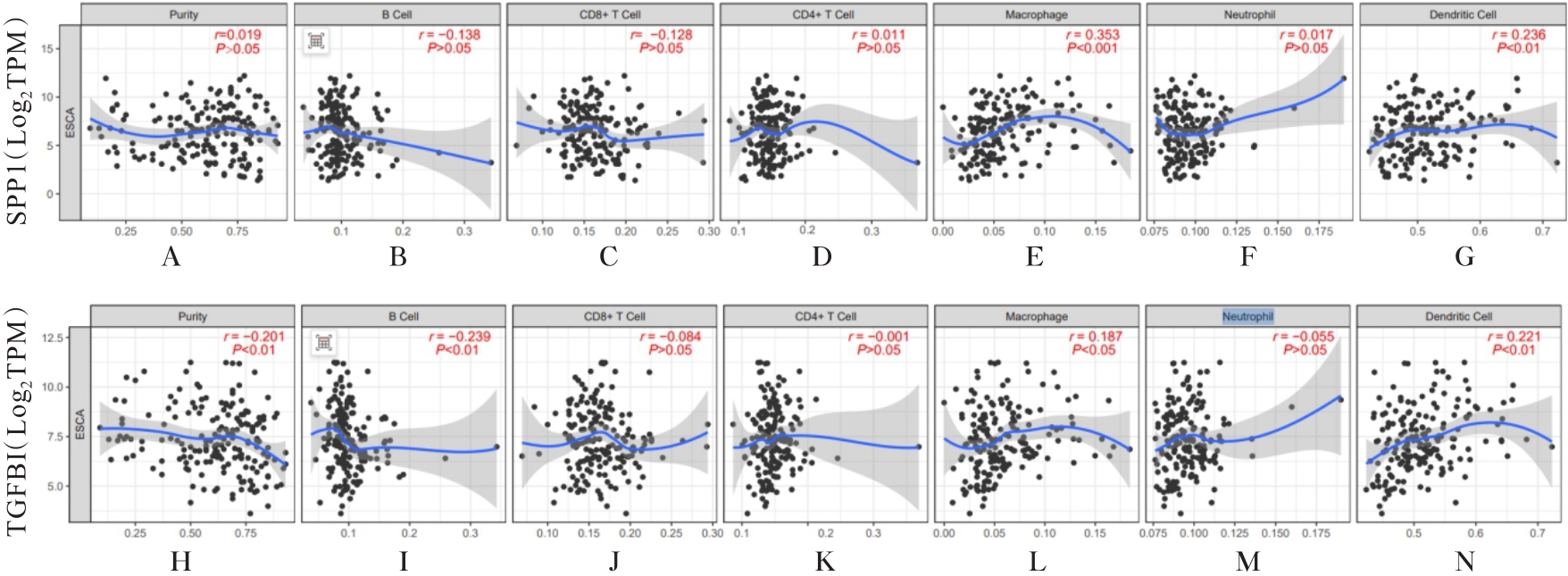
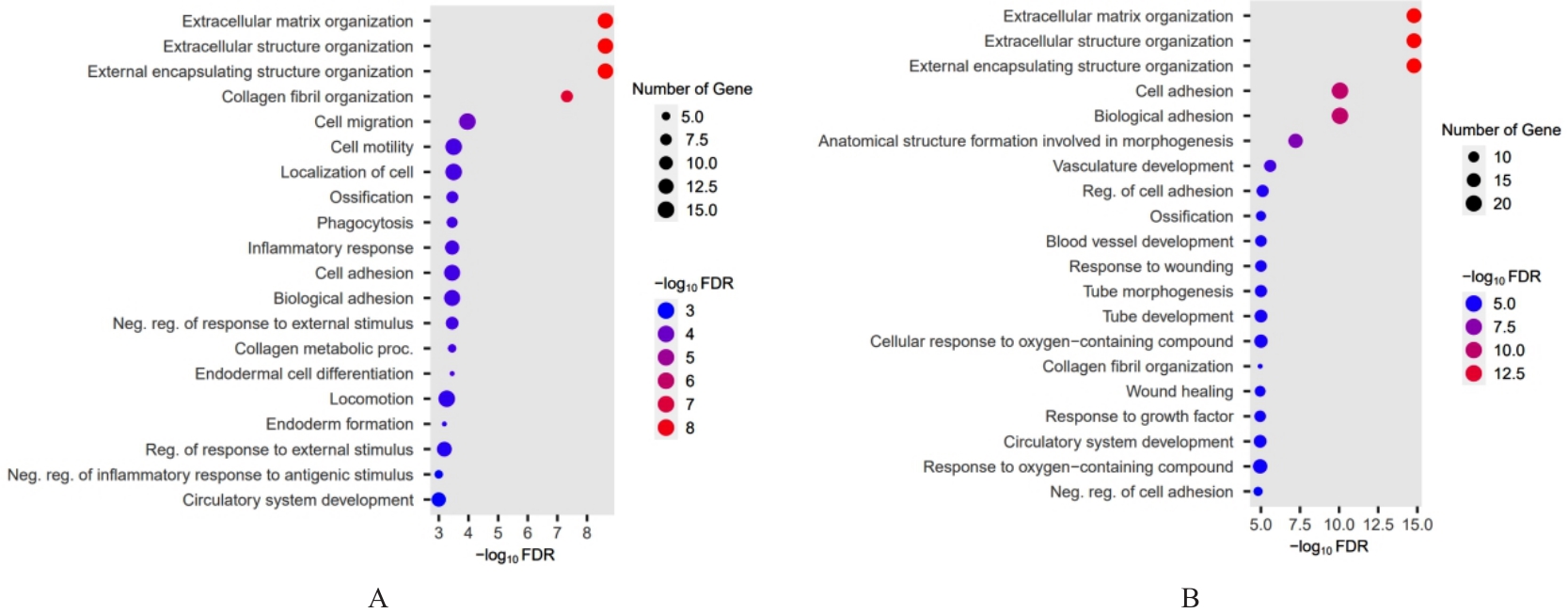
Bioinformatics analysis based on relationship between SSP1 and TGFB1 and occurrence, prognosis, and immune invasion of esophageal adenocarcinoma
- Department of Cardio-Thoracic Surgery,General Hospital,Tianjin Medical University,Tianjin 300052,China
-
Received:2023-03-22Online:2024-07-28Published:2024-08-01 -
Contact:Peng ZHANG E-mail:zhangfapeng@aliyun.com
CLC Number:
- R735.1
Cite this article
Yuanguo WANG,Peng ZHANG. Bioinformatics analysis based on relationship between SSP1 and TGFB1 and occurrence, prognosis, and immune invasion of esophageal adenocarcinoma[J].Journal of Jilin University(Medicine Edition), 2024, 50(4): 1076-1086.
share this article
Tab. 1
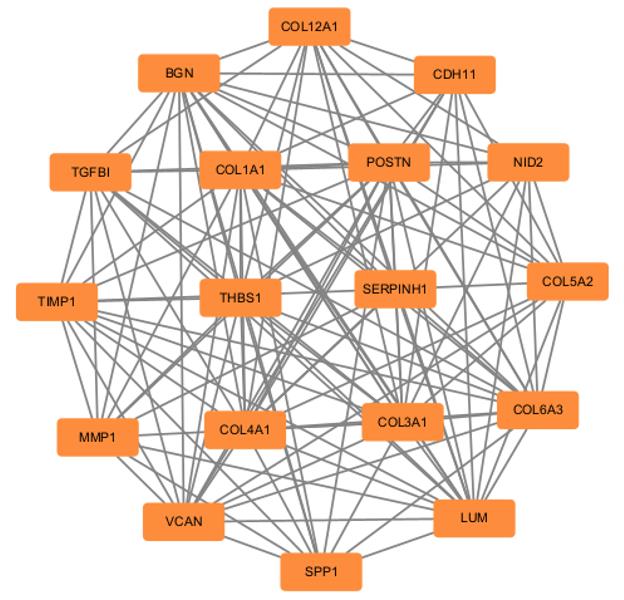
KEGG signaling pathway enrichment analysis on SPP1 positively related genes"
| KEGG pathway | Count | P | Gene |
|---|---|---|---|
| Protein digestion and absorption | 8 | <0.01 | COL1A1, COL3A1, COL1A2, COL5A1, COL24A1, COL11A1, COL7A1, COL10A1 |
| AGE-RAGE signaling pathway in diabetic complications | 6 | <0.01 | COL1A1, COL3A1, COL1A2, FN1, NOX4, MAPK12 |
| Osteoclast differentiation | 6 | <0.01 | FCGR3A, FCGR2A, NCF2, TREM2, FCGR1A, MAPK12 |
| Leishmaniasis | 5 | <0.01 | FCGR3A, FCGR2A, NCF2, FCGR1A, MAPK12 |
| Neutrophil extracellular trap formation | 6 | <0.01 | FCGR3A, FCGR2A, SIGLEC9, NCF2, FCGR1A, MAPK12 |
| Platelet activation | 5 | <0.01 | COL1A1, COL3A1, FCGR2A, COL1A2, MAPK12 |
| Phagosome | 5 | <0.01 | MSR1, FCGR3A, FCGR2A, NCF2, FCGR1A |
| ECM-receptor interaction | 4 | <0.01 | COL1A1, COL1A2, SPP1, FN1 |
| Fc gamma R-mediated phagocytosis | 4 | <0.01 | HCK, FCGR3A, FCGR2A, FCGR1A |
| Diabetic cardiomyopathy | 5 | <0.01 | COL1A1, COL3A1, COL1A2, NCF2, MAPK12 |
| Proteoglycans in cancer | 5 | <0.01 | COL1A1, COL1A2, PLAU, FN1, MAPK12 |
| Amoebiasis | 4 | <0.01 | COL1A1, COL3A1, COL1A2, FN1 |
| Relaxin signaling pathway | 4 | <0.01 | COL1A1, COL3A1, COL1A2, MAPK12 |
| Tuberculosis | 4 | <0.05 | FCGR3A, FCGR2A, FCGR1A, MAPK12 |
| Focal adhesion | 4 | <0.05 | COL1A1, COL1A2, SPP1, FN1 |
| Staphylococcus aureus infection | 3 | <0.05 | FCGR3A, FCGR2A, FCGR1A |
Tab.2
KEGG signaling pathway enrichment analysis on TGFB1 positively related genes"
| KEGG pathway | Count | P | Gene |
|---|---|---|---|
| Focal adhesion | 8 | <0.01 | COL1A1, CAV2, ACTN1, CAV1, COL6A2, TNC, ITGA5, PGF |
| Proteoglycans in cancer | 6 | <0.01 | COL1A1, TGFB1, PLAU, CAV2, CAV1, ITGA5 |
| ECM-receptor interaction | 4 | <0.01 | COL1A1, COL6A2, TNC, ITGA5 |
| Protein digestion and absorption | 4 | <0.01 | COL1A1, COL6A2, COL5A3, COL7A1 |
| PI3K-AKT signaling pathway | 5 | <0.05 | COL1A1, COL6A2, TNC, ITGA5, PGF |
| Bacterial invasion of epithelial cells | 3 | <0.05 | CAV2, CAV1, ITGA5 |
| AGE-RAGE signaling pathway in diabetic complications | 3 | <0.05 | COL1A1, TGFB1, SERPINE1 |
| Amoebiasis | 3 | <0.05 | COL1A1, TGFB1, ACTN1 |
| 1 | COLEMAN H G, XIE S H, LAGERGREN J. The epidemiology of esophageal adenocarcinoma[J]. Gastroenterology, 2018, 154(2): 390-405. |
| 2 | POLEBOYINA P K, ALAGUMUTHU M, PASHA A, et al. Entrectinib a plausible inhibitor for osteopontin (SPP1) in cervical cancer-integrated bioinformatic approach[J]. Appl Biochem Biotechnol, 2023, 195(12): 7766-7795. |
| 3 | ZHAO K D, MA Z, ZHANG W. Comprehensive analysis to identify SPP1 as a prognostic biomarker in cervical cancer[J]. Front Genet, 2022, 12: 732822. |
| 4 | XIE W, CHENG J, HONG Z J, et al. Multi-transcriptomic analysis reveals the heterogeneity and tumor-promoting role of SPP1/CD44-mediated intratumoral crosstalk in gastric cancer[J]. Cancers, 2022, 15(1): 164. |
| 5 | STREEL G D, LUCAS S. Targeting immunosuppression by TGF-β1 for cancer immunotherapy[J]. Biochem Pharmacol, 2021, 192: 114697. |
| 6 | SOUKUPOVA J, MALFETTONE A, BERTRAN E, et al. Epithelial-mesenchymal transition (EMT) induced by TGF-β in hepatocellular carcinoma cells reprograms lipid metabolism[J]. Int J Mol Sci, 2021, 22(11): 5543. |
| 7 | MOREAU J M, VELEGRAKI M, BOLYARD C, et al. Transforming growth factor-β1 in regulatory T cell biology[J]. Sci Immunol, 2022, 7(69): eabi4613. |
| 8 | 田 东, 付茂勇, 赵泽良, 等. 食管腺癌临床病理特征及影响预后的因素[J]. 现代肿瘤医学, 2013, 21(8): 1748-1753. |
| 9 | RUSTGI A K, EL-SERAG H B. Esophageal carcinoma[J]. N Engl J Med, 2014, 371(26): 2499-2509. |
| 10 | RUBENSTEIN J H, TAYLOR J B. Meta-analysis: the association of oesophageal adenocarcinoma with symptoms of gastro-oesophageal reflux[J]. Aliment Pharmacol Ther, 2010, 32(10): 1222-1227. |
| 11 | SNIDER E J, FREEDBERG D E, ABRAMS J A. Potential role of the microbiome in barrett’s esophagus and esophageal adenocarcinoma[J]. Dig Dis Sci, 2016, 61(8): 2217-2225. |
| 12 | KHALAFI S, LOCKHART A C, LIVINGSTONE A S, et al. Targeted molecular therapies in the treatment of esophageal adenocarcinoma, are we there yet?[J]. Cancers, 2020, 12(11): 3077. |
| 13 | ZHANG X, WANG Y X, MENG L H. Comparative genomic analysis of esophageal squamous cell carcinoma and adenocarcinoma: new opportunities towards molecularly targeted therapy[J]. Acta Pharm Sin B, 2022, 12(3): 1054-1067. |
| 14 | FATEHI HASSANABAD A, CHEHADE R, BREADNER D, et al. Esophageal carcinoma: towards targeted therapies[J]. Cell Oncol, 2020, 43(2): 195-209. |
| 15 | 刘 迁, 祁国萍, 于华裔, 等. 结肠癌核心基因和独立预后因子筛选的生物信息学分析[J]. 吉林大学学报(医学版), 2022, 48(3): 755-765. |
| 16 | MESSEX J K, BYRD C J, THOMAS M U, et al. Macrophages cytokine Spp1 increases growth of prostate intraepithelial neoplasia to promote prostate tumor progression[J]. Int J Mol Sci, 2022, 23(8): 4247. |
| 17 | GÖTHLIN EREMO A, LAGERGREN K, OTHMAN L, et al. Evaluation of SPP1/osteopontin expression as predictor of recurrence in tamoxifen treated breast cancer[J]. Sci Rep, 2020, 10(1): 1451. |
| 18 | ZHANG W G, FAN J L, CHEN Q, et al. SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma[J]. Oncol Lett, 2018, 15(5): 7028-7036. |
| 19 | PERIYASAMY A, GOPISETTY G, SUBRAMANIUM M J, et al. Identification and validation of differential plasma proteins levels in epithelial ovarian cancer[J]. J Proteomics, 2020, 226: 103893. |
| 20 | ZENG B, ZHOU M, WU H, et al. SPP1 promotes ovarian cancer progression via Integrin β1/FAK/AKT signaling pathway[J]. Onco Targets Ther, 2018, 11: 1333-1343. |
| 21 | LIU L L, ZHANG R Y, DENG J W, et al. Construction of TME and Identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma[J]. Cancer Immunol Immunother, 2022, 71(1): 121-136. |
| 22 | PANG X C, ZHANG J L, HE X, et al. SPP1 promotes enzalutamide resistance and epithelial-mesenchymal-transition activation in castration-resistant prostate cancer via PI3K/AKT and ERK1/2 pathways[J]. Oxid Med Cell Longev, 2021, 2021: 5806602. |
| 23 | RABJERG M, BJERREGAARD H, HALEKOH U, et al. Molecular characterization of clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1 as promising novel prognostic markers[J]. APMIS, 2016, 124(5): 372-383. |
| 24 | CHEN J W, HOU C X, ZHENG Z T, et al. Identification of secreted phosphoprotein 1 (SPP1) as a prognostic factor in lower-grade gliomas[J]. World Neurosurg, 2019, 130: e775-e785. |
| 25 | BIE T W, ZHANG X W. Higher expression of SPP1 predicts poorer survival outcomes in head and neck cancer[J]. J Immunol Res, 2021, 2021: 8569575. |
| 26 | LIU Y H, ZHANG L, JU X Y, et al. Single-cell transcriptomic analysis reveals macrophage-tumor crosstalk in hepatocellular carcinoma[J]. Front Immunol, 2022, 13: 955390. |
| 27 | GAO W, LIU D L, SUN H Y, et al. SPP1 isa prognostic related biomarker and correlated with tumor-infiltrating immune cells in ovarian cancer[J]. BMC Cancer, 2022, 22(1): 1367. |
| 28 | GUASCH G, SCHOBER M, PASOLLI H A, et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia[J]. Cancer Cell, 2007, 12(4): 313-327. |
| 29 | SYED V. TGF-β signaling in cancer[J]. J Cell Biochem, 2016, 117(6): 1279-1287. |
| 30 | IVANOV S V, IVANOVA A V, SALNIKOW K, et al. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors[J]. Biochem Biophys Res Commun, 2008, 370(4): 536-540. |
| 31 | SHANG D H, LIU Y T, YANG P Q, et al. TGFBI-promoted adhesion, migration and invasion of human renal cell carcinoma depends on inactivation of von Hippel-Lindau tumor suppressor[J]. Urology, 2012, 79(4): 966.e1-966.e7. |
| 32 | FICO F, SANTAMARIA-MARTÍNEZ A. TGFBI modulates tumour hypoxia and promotes breast cancer metastasis[J]. Mol Oncol, 2020, 14(12): 3198-3210. |
| 33 | HAN B, CAI H L, CHEN Y, et al. The role of TGFBI (βig-H3) in gastrointestinal tract tumorigenesis[J]. Mol Cancer, 2015, 14: 64. |
| 34 | WEN G Y, PARTRIDGE M A, LI B Y, et al. TGFBI expression reduces in vitro and in vivo metastatic potential of lung and breast tumor cells[J]. Cancer Lett, 2011, 308(1): 23-32. |
| [1] | Chaojie GUO,Jiajia ZHANG,Jie ZENG,Huiyu WANG, AIERFATI·Aimaier,Jiang XU. Expressions of PLOD1 in oral squamous cell carcinoma tissue and cells and their significances [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 1035-1043. |
| [2] | Jinlian LI,Lanzhen HUANG,Xishi HUANG,Kangzhi LI,Jiali JIANG,Miaomiao ZHANG,Qunying WU. Bioinformatics analysis on key genes related to prognosis, diagnosis, and immune cell infiltration of hepatocellular carcinoma and their potential therapeutic drugs [J]. Journal of Jilin University(Medicine Edition), 2024, 50(4): 1062-1075. |
| [3] | Linghui LIN,Na LI,Xiaoyan YIN,Xiaoling WANG,Yaping HU,Wei LIU,Rui FEI,Xinli TIAN. Bioinformatics anlysis based on three-dimensional structure of Helicobacter pylori hp0169 gene [J]. Journal of Jilin University(Medicine Edition), 2024, 50(3): 739-748. |
| [4] | Zijia ZHU,Xia CHEN,Man CUI,Jihong WEN,Ping WANG,Dong SONG. Bioinformatics and molecular docking technology analysis on mechanism of salidroside on key differential genes of triple negative breast cancer [J]. Journal of Jilin University(Medicine Edition), 2024, 50(3): 759-769. |
| [5] | Yuting LIU,Ying YU,Guizhen LI,Qinxue SHI,Binbin LI. Bioinformatics analysis based on expression of splicing factor SRSF9 in head and neck squamous cell carcinoma and clinical significance [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 379-391. |
| [6] | Yueying SONG,Chao GAO,Wenjun CHEN,Aiyu SHAO,Yichun QIAO,Zhuolin LI. Screening of miRNAs related to prognosis of triple-negative breast cancer and its gene network based on TCGA Database [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 392-399. |
| [7] | Liping CHEN,Li HAN,Hua BIAN,Liye PANG. Bioinformatics analysis based on differentially expressed genes and screening of traditional Chinese medicine for treatment of severe bronchial asthma [J]. Journal of Jilin University(Medicine Edition), 2024, 50(2): 411-421. |
| [8] | Minqi NING,Yong HE,Bingshu LI,Guotao HUANG,Xiaohu ZUO,Zhihan ZHAO,Wuyue HAN,Li HONG. Bioinformatics analysis based on pelvic organ prolapse related aging genes of GEO Database and LASSO regression algorithm [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 178-187. |
| [9] | Zixu YANG,Chang SU,Boyuan WANG,Chong LIU,Minghe LI. Bioinformatics analysis on molecular subtypes and clinical characteristics of head and neck squamous cell carcinoma based on genes associated with lactate metabolism [J]. Journal of Jilin University(Medicine Edition), 2024, 50(1): 198-207. |
| [10] | Yaqi XU,Yanyu WANG,Wenjing ZHANG,Mei HAN,Huaxia MU,Xi YANG,Weixiao BU,Zikun TAO,Yujia KONG,Fuyan SHI,Suzhen WANG. Bioinformatics analysis on screening of key genes of hepatitis B virus-related hepatocellular carcinoma and its relationship with prognosis [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1243-1252. |
| [11] | Haikang CUI,Xudong ZHANG,Xiaoning LI,Xi YANG,Lan YANG,Wenjie ZHANG. Bioinformatics analysis on predition effect of subtypes of cell pyroptosis and APOD on prognosis of gastric cancer patients [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1268-1279. |
| [12] | Yiming ZHAO,Haiyang XU. Bioinformatics analysis on related genes and candidate pathways of glioblastoma multiforme [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1280-1289. |
| [13] | Xiaoying ZHAO,Jiansheng SU,Jiahui MA,Yuefeng WANG,Dan WANG,Liyuan SUN. Bioinformatics analysis based on activity of heterotrimeric peptide H5LL and construction of recombinant Lactic acid bacteria expression vector [J]. Journal of Jilin University(Medicine Edition), 2023, 49(5): 1366-1374. |
| [14] | Jingjing LI, REZIWANGULI·Aisikaier,Wanyi XING,Yinggang ZOU. Primary dedifferentiated liposarcoma of unilateral ovary: A case report and literature review [J]. Journal of Jilin University(Medicine Edition), 2023, 49(4): 1040-1045. |
| [15] | Renyi YANG,Shuwang PENG,Yongheng WANG,Yuxuan DONG,Shanshan DUAN. Construction of ferroptosis prognostic risk model of thyroid cancer and bioinformatics analysis on its potential mechanism [J]. Journal of Jilin University(Medicine Edition), 2023, 49(2): 402-413. |