吉林大学学报(工学版) ›› 2025, Vol. 55 ›› Issue (1): 297-306.doi: 10.13229/j.cnki.jdxbgxb.20230267
• 计算机科学与技术 • 上一篇
基于深度学习的核糖核酸二级结构预测方法
- 1.吉林大学 计算机科学与技术学院,长春 130012
2.吉林大学 符号计算与知识工程教育部重点实验室,长春 130012
3.长崎综合科学大学 研究生院工学研究科,长崎 851-0193
Deep learning-based method for ribonucleic acid secondary structure prediction
Yuan-ning LIU1,2( ),Zi-nan ZANG1,2,Hao ZHANG1,2(
),Zi-nan ZANG1,2,Hao ZHANG1,2( ),Zhen LIU1,3
),Zhen LIU1,3
- 1.College of Computer Science and Technology,Jilin University,Changchun 130012,China
2.Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education,Jilin University,Changchun 130012,China
3.Graduate School of Engineering,Nagasaki Institute of Applied Science,Nagasaki 851-0193,Japan
摘要:
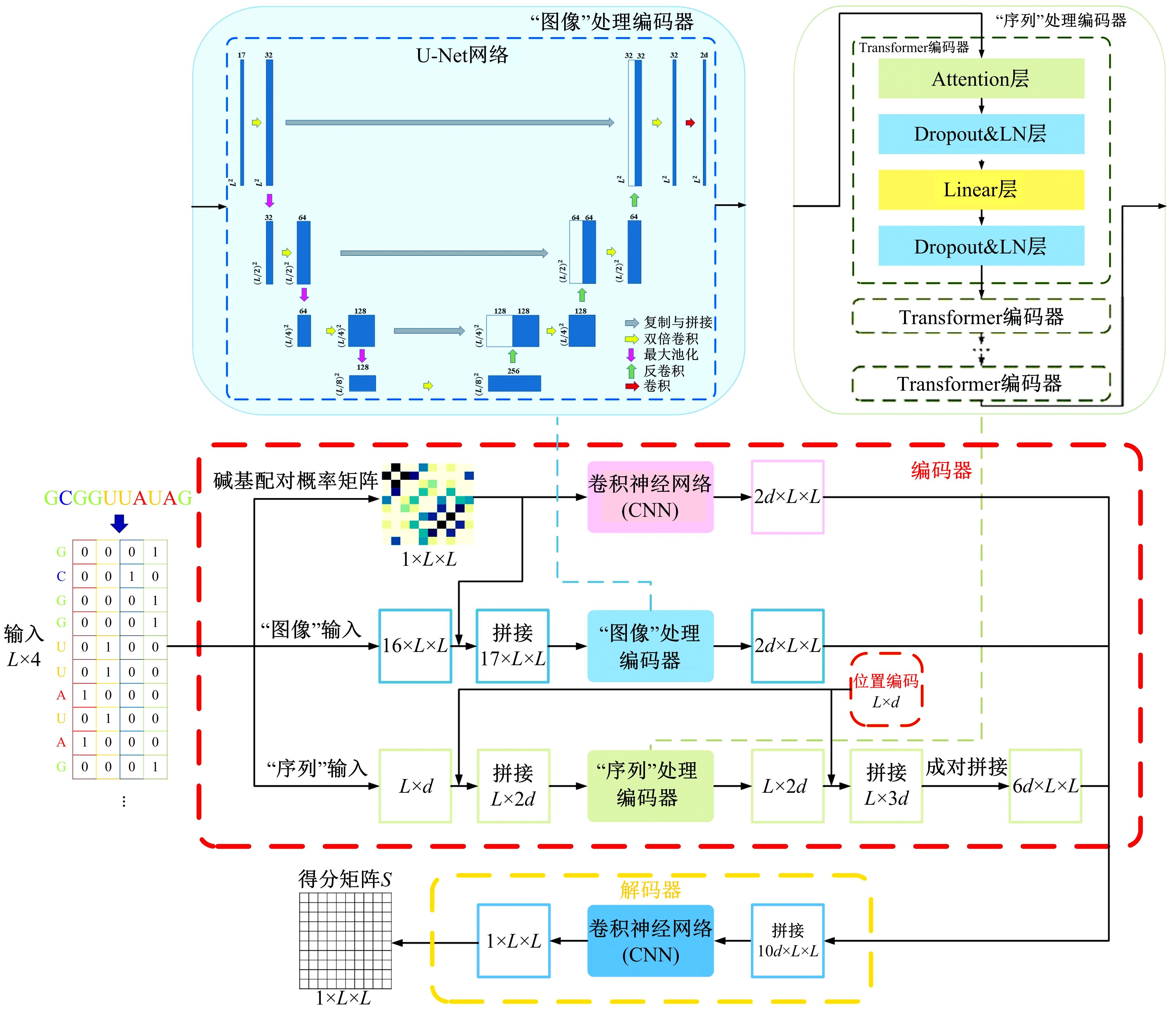
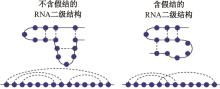
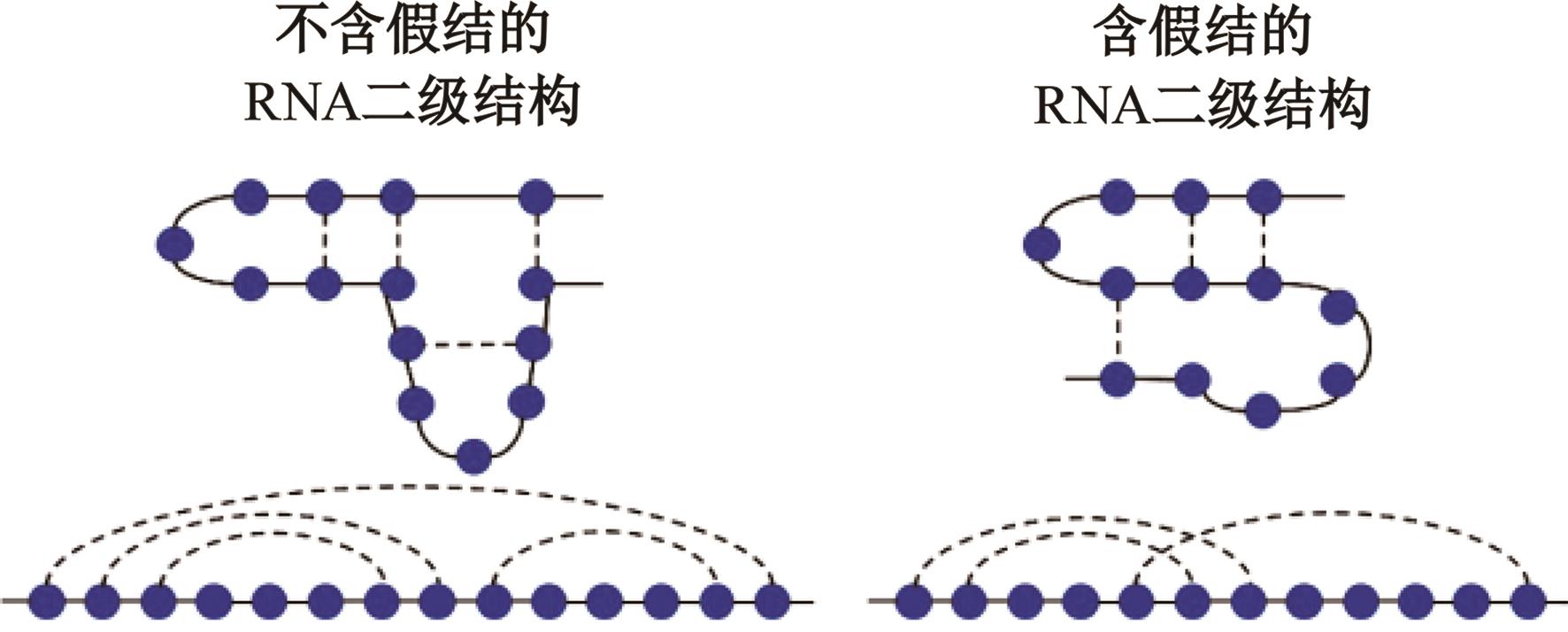
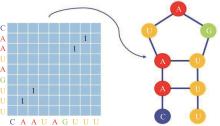
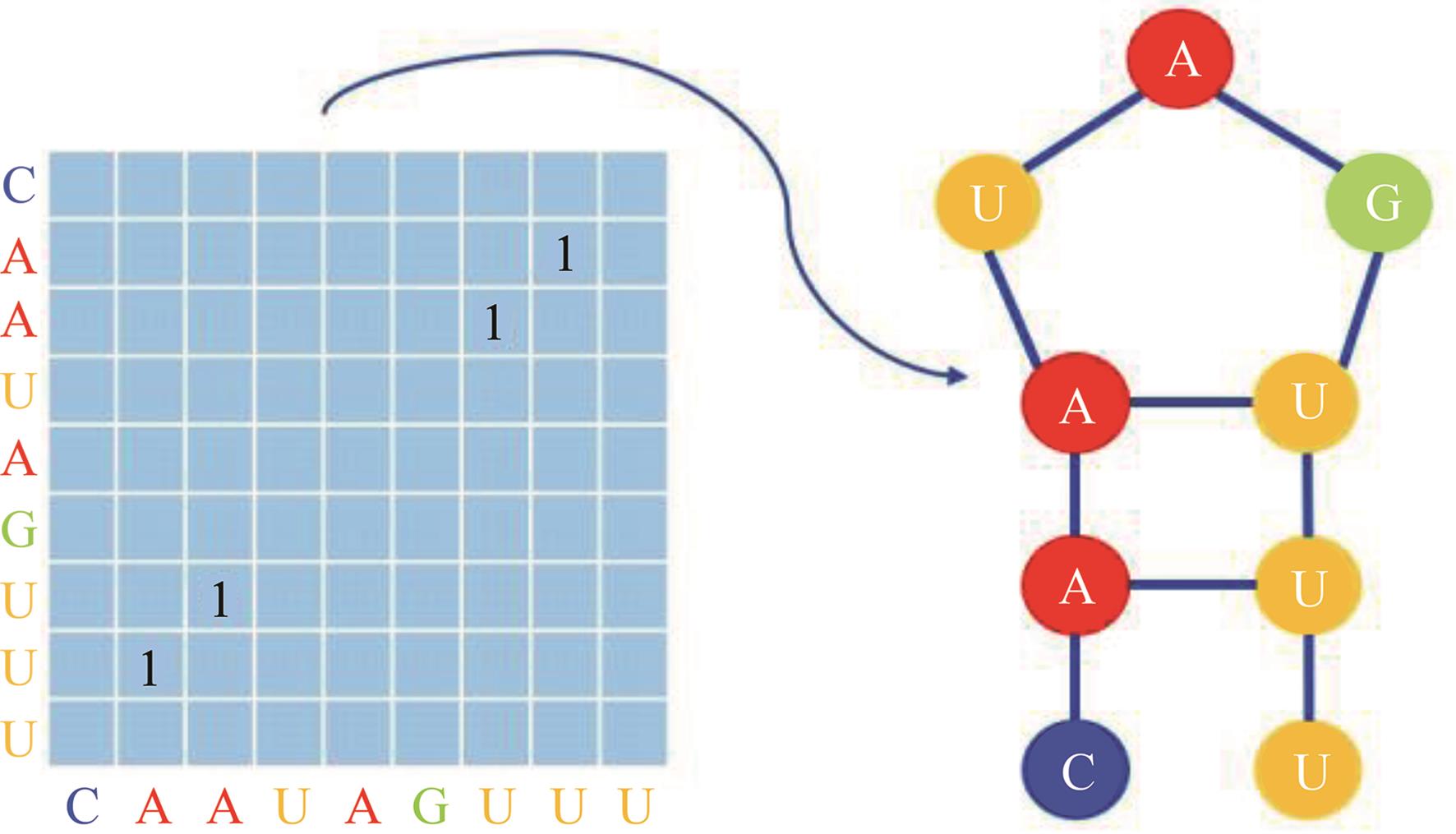
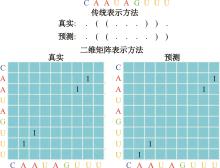
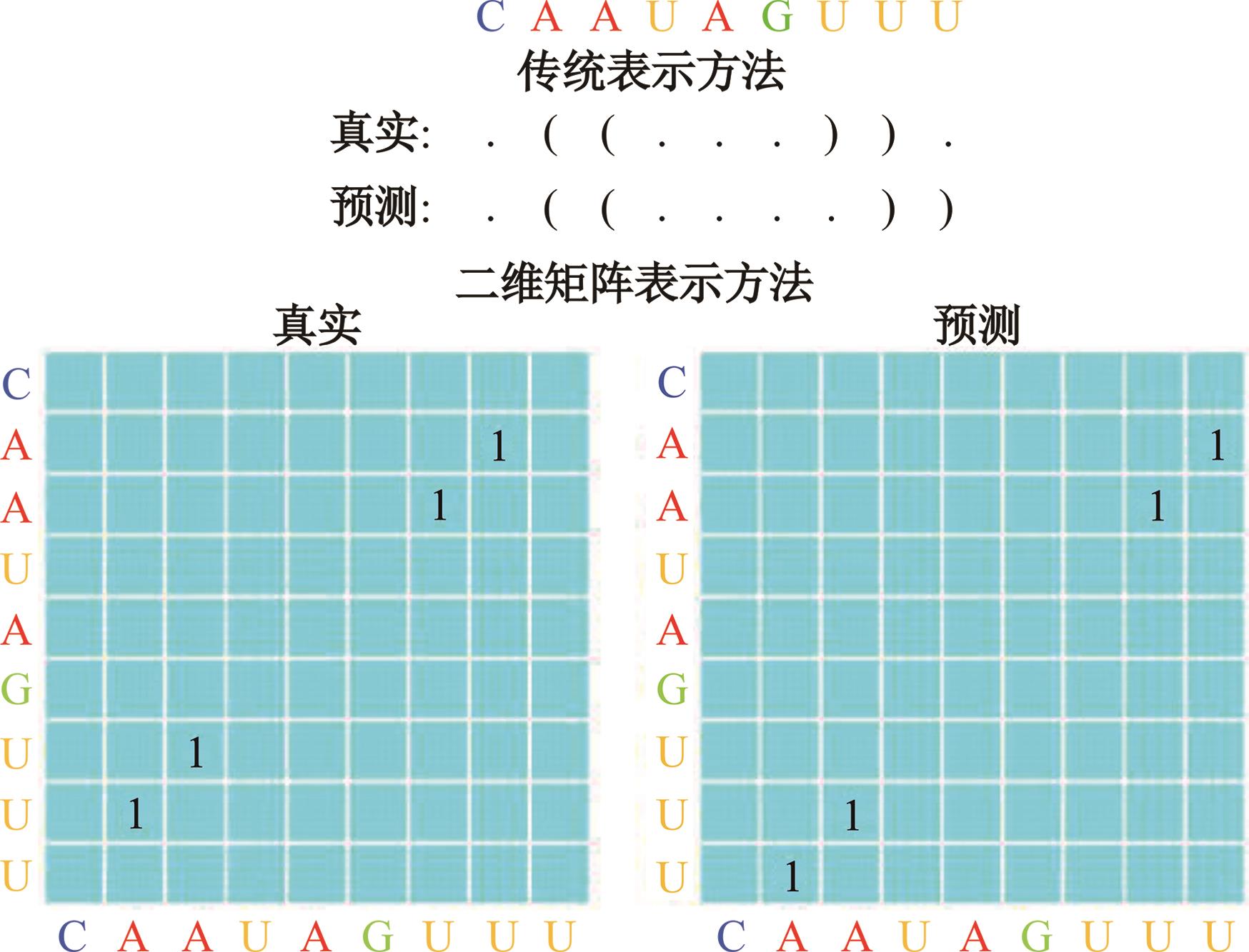
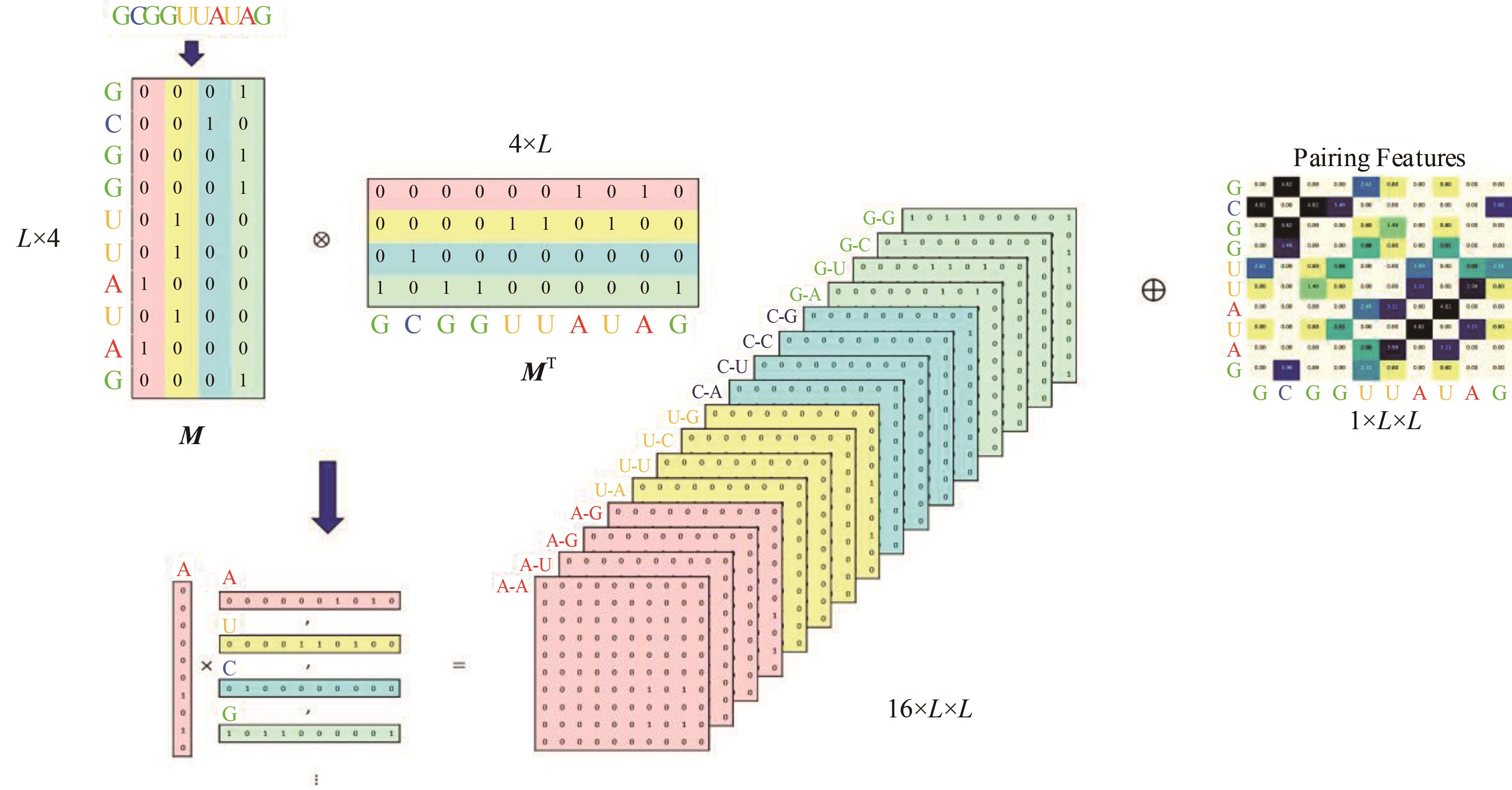
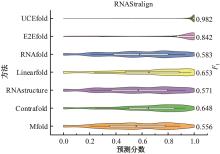
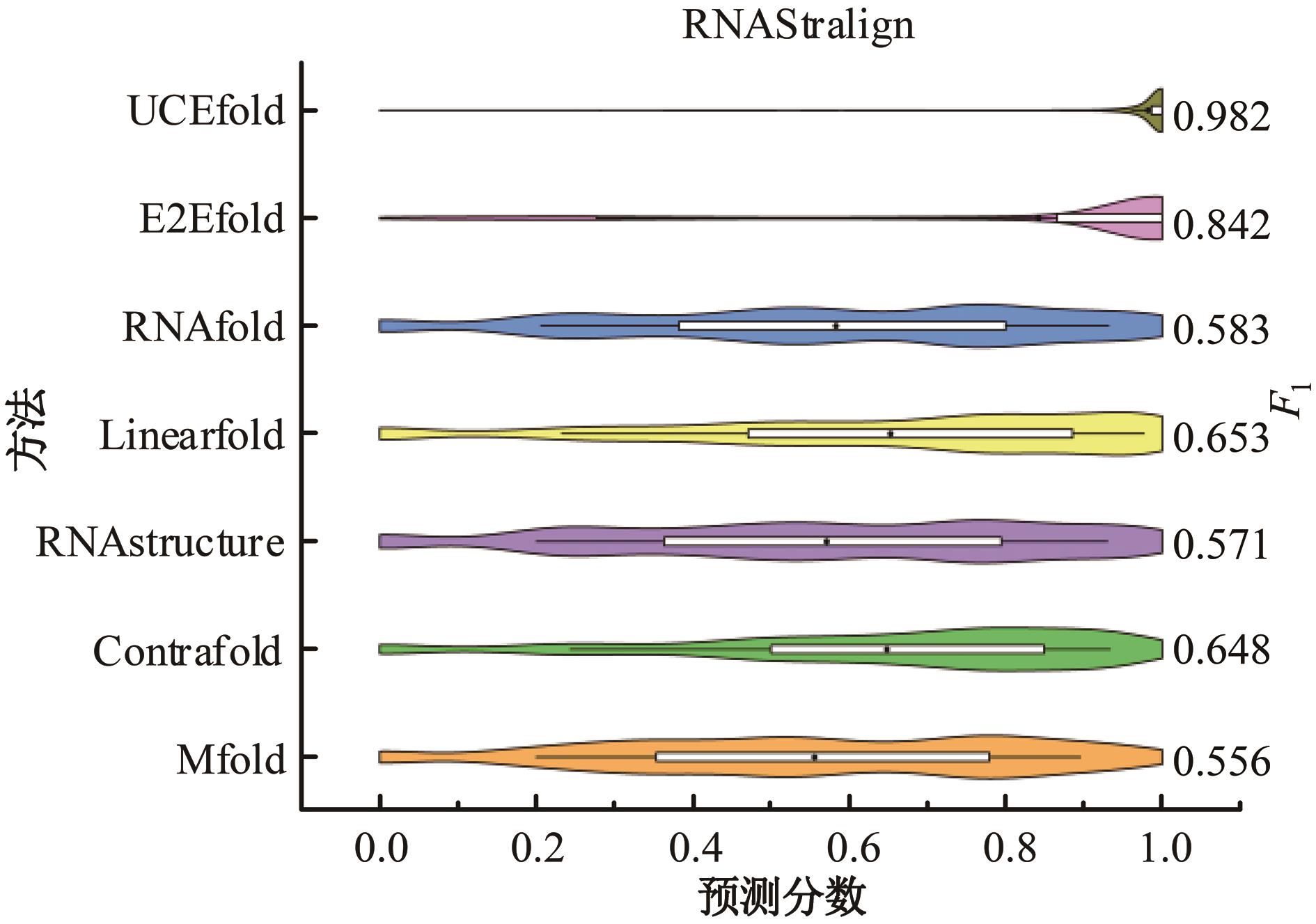
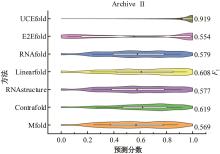
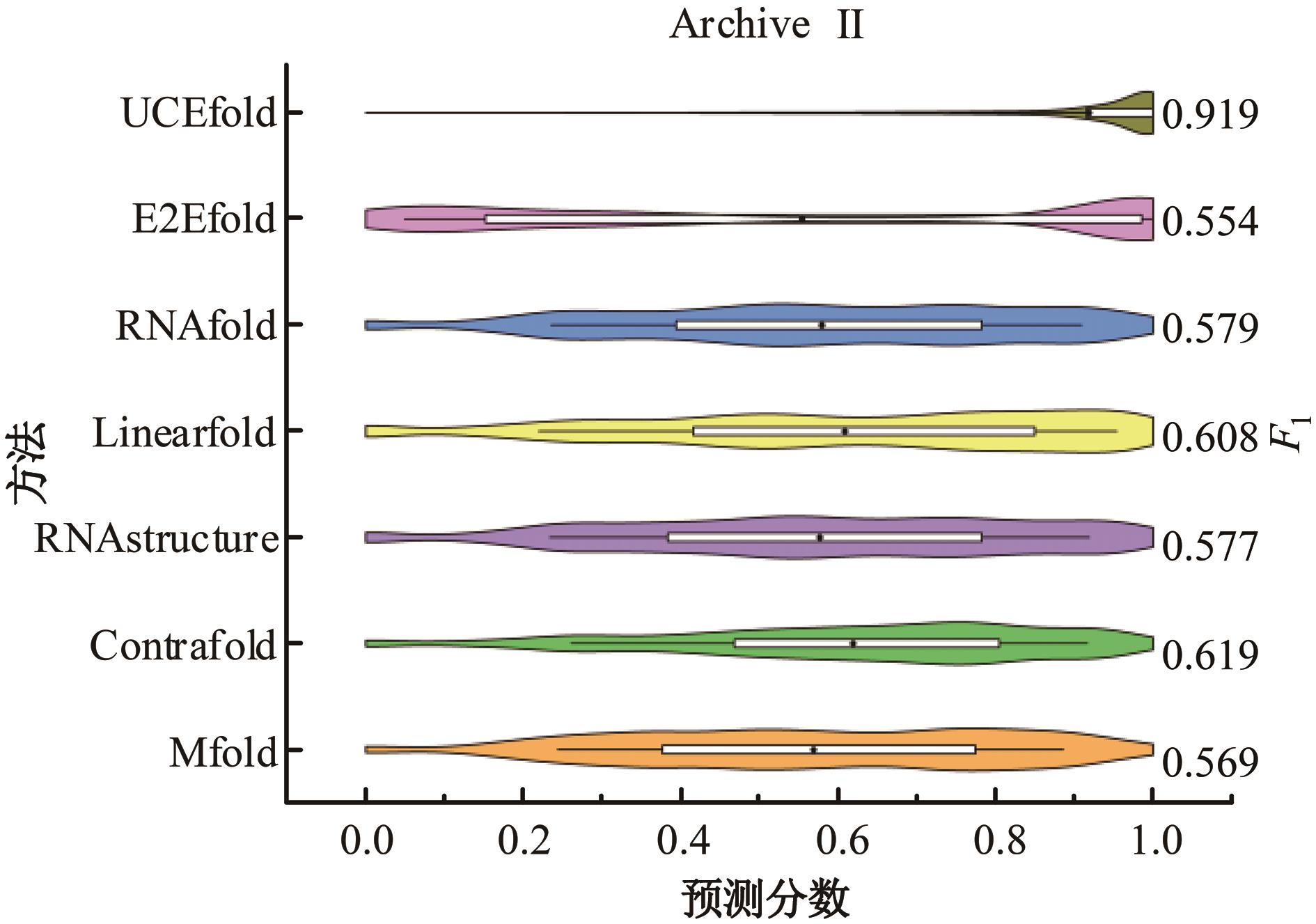
本文提出了一种基于深度学习的方法UCEfold,用于预测核糖核酸(Ribonucleic acid,RNA)二级结构。UCEfold是一种同时采用“序列”和“图像”作为深度学习模型输入提取隐藏特征的全新方法,并在模型中加入一定的先验知识提高预测精度。在RNAStralign和ArchiveⅡ两个数据集上测试UCEfold模型,结果表明UCEfold性能显著优于传统方法,能够更准确地预测带假结的RNA序列,并具有较强的泛化能力,有效解决了传统算法复杂度高、效率低下且无法预测假结的瓶颈。
中图分类号:
- TP399
| 1 | Crick F. Central dogma of molecular biology[J]. Nature, 1970, 227: 561-563. |
| 2 | Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription[J]. Science, 2007, 316: 1484-1488. |
| 3 | Sharp P. The centrality of RNA[J]. Cell, 2009, 136: 577-580. |
| 4 | Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction[J]. Nucleic Acids Research, 2003, 31: 3406-3415. |
| 5 | Lorenz R, Bernhart S H, Höner Zu Siederdissen C, et al. ViennaRNA Package 2.0[J]. Algorithms for Molecular Biology, 2011, 6: No.26. |
| 6 | Mathews D H, Turner D H. Prediction of RNA secondary structure by free energy minimization[J]. Current Opinion in Structural Biology, 2006, 16: 270-278. |
| 7 | Huang L, Zhang H, Deng D, et al. LinearFold: linear-time approximate RNA folding by 5'-to-3' dynamic programming and beam search[J]. Bioinformatics, 2019, 35: i295-i304. |
| 8 | Brierley I, Pennell S, Gilbert R J C. Viral RNA pseudoknots: versatile motifs in gene expression and replication[J]. Nature Reviews Microbiology, 2007, 5: 598-610. |
| 9 | Bernhart S H, Hofacker I L, Will S, et al. RNAalifold: improved consensus structure prediction for RNA alignments[J]. BMC Bioinformatics, 2008, 9: No.474. |
| 10 | Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars[J]. Nucleic Acids Research, 2003, 31: 3423-3428. |
| 11 | Do C B, Woods D A, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models[J]. Bioinformatics, 2006, 22: e90-e98. |
| 12 | Zakov S, Goldberg Y, Elhadad M, et al. Rich parameterization improves RNA structure prediction. [J]. Journal of Computational Biology: A Journal of Computational Molecular Cell Biology, 2011, 6577: 546-562. |
| 13 | Zhang H, Zhang C, Li Z, et al. A new method of RNA secondary structure prediction based on convolutional neural network and dynamic programming[J]. Frontiers in Genetics, 2019, 10: No.467 |
| 14 | Chen X, Li Y, Umarov R, et al. RNA secondary structure prediction by learning unrolled algorithms [C]∥Proceedings of the International Conference on Learning Representations(ICLR), Addis Ababa, Ethiopia, 2020: 1-19. |
| 15 | Sato K, Akiyama M, Sakakibara Y. RNA secondary structure prediction using deep learning with thermodynamic integration[J]. Nature Communications, 2021, 12(1): No. 941. |
| 16 | Singh J, Hanson J, Paliwal K, et al. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning[J]. Nature Communications, 2019, 10(1): No. 5407. |
| 17 | Tan Z, Fu Y, Sharma G, et al. TurboFold Ⅱ: RNA structural alignment and secondary structure prediction informed by multiple homologs[J]. Nucleic Acids Research, 2017, 45(20): 11570-11581. |
| 18 | Sloma M, Mathews D. Exact calculation of loop formation probability identifies folding motifs in RNA secondary structures[J]. RNA, 2016, 22(12): 1808-1818. |
| 19 | Hochreiter S, Schmidhuber J. Long short-term memory[J]. Neural Computation, 1997, 9: 1735-1780. |
| 20 | Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need[C]∥Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, USA, 2017: 6000-6010. |
| 21 | Fu L, Cao Y, Wu J, et al. UFold: fast and accurate RNA secondary structure prediction with deep learning[J]. Nucleic Acids Research, 2022, 50(3): No.e14. |
| 22 | Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation [C]∥Proceedings of the Medical Image Computing and Computer-Assisted Intervention(MICCAI), Munich, Germany, 2015: 234-241. |
| [1] | 王勇,边宇霄,李新潮,徐椿明,彭刚,王继奎. 基于多尺度编码-解码神经网络的图像去雾算法[J]. 吉林大学学报(工学版), 2024, 54(12): 3626-3636. |
| [2] | 王清永,曲伟强. 基于线性规划的城市轨道交通运行调度优化算法[J]. 吉林大学学报(工学版), 2023, 53(12): 3446-3451. |
| [3] | 高海龙,徐一博,侯德藻,王雪松. 基于深度异步残差网络的路网短时交通流预测算法[J]. 吉林大学学报(工学版), 2023, 53(12): 3458-3464. |
| [4] | 王军,王华琳,黄博文,付强,刘俊. 基于联邦学习和自注意力的工业物联网入侵检测[J]. 吉林大学学报(工学版), 2023, 53(11): 3229-3237. |
| [5] | 周丰丰,颜振炜. 基于混合特征的特征选择神经肽预测模型[J]. 吉林大学学报(工学版), 2023, 53(11): 3238-3245. |
| [6] | 孙舒杨,程玮斌,张浩桢,邓向萍,齐红. 基于深度学习的两阶段实时显式拓扑优化方法[J]. 吉林大学学报(工学版), 2023, 53(10): 2942-2951. |
| [7] | 王生生,李晨旭,王翔宇,姚志林,刘一申,吴佳倩,杨晴然. 基于改进残差胶囊网络和麻雀搜索的脑瘤图像分类[J]. 吉林大学学报(工学版), 2022, 52(11): 2653-2661. |
| [8] | 周丰丰,张亦弛. 基于稀疏自编码器的无监督特征工程算法BioSAE[J]. 吉林大学学报(工学版), 2022, 52(7): 1645-1656. |
| [9] | 魏晓辉,苗艳微,王兴旺. Rhombus sketch:自适应和准确的流数据sketch[J]. 吉林大学学报(工学版), 2022, 52(4): 874-884. |
| [10] | 刘桂霞,裴志尧,宋佳智. 基于深度学习的蛋白质⁃ATP结合位点预测[J]. 吉林大学学报(工学版), 2022, 52(1): 187-194. |
| [11] | 宋荷庆,尤力强,宋元,王章野. 面向云端系统的可伸缩群体远程对外证明方法[J]. 吉林大学学报(工学版), 2021, 51(6): 2198-2206. |
| [12] | 董延华,刘靓葳,赵靖华,李亮,解方喜. 基于BPNN在线学习预测模型的扭矩实时跟踪控制[J]. 吉林大学学报(工学版), 2021, 51(4): 1405-1413. |
| [13] | 魏晓辉,汤钫宇,李洪亮. 地理分布数据中心的工作流经济高效资源分配[J]. 吉林大学学报(工学版), 2021, 51(4): 1349-1357. |
| [14] | 魏晓辉,周长宝,沈笑先,刘圆圆,童群超. 机器学习加速CALYPSO结构预测的可行性[J]. 吉林大学学报(工学版), 2021, 51(2): 667-676. |
| [15] | 陈蔓,钟勇,李振东. 隐低秩结合低秩表示的多聚焦图像融合[J]. 吉林大学学报(工学版), 2020, 50(1): 297-305. |
|
||